Joe Granger Methionine Repressor: Escherichia coli
From Proteopedia
| |||||||||
| 1cma, resolution 2.80Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
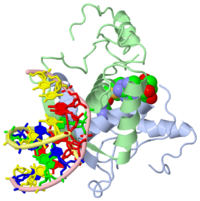
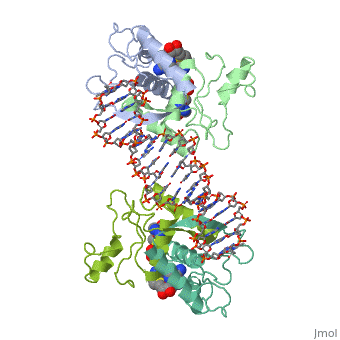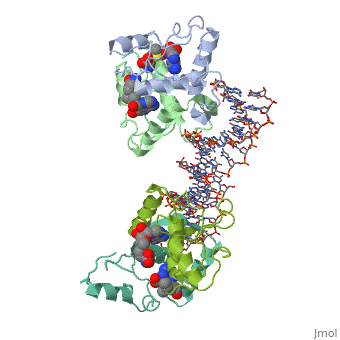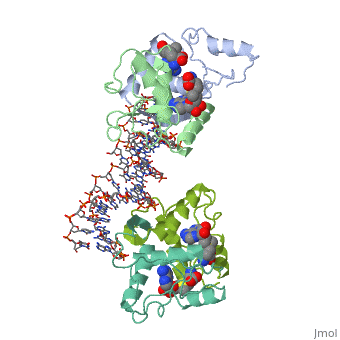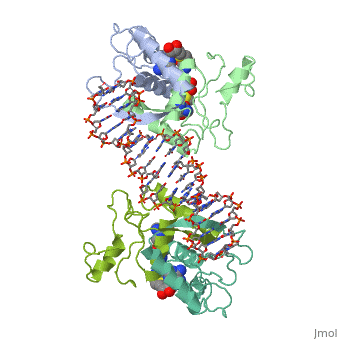
MET REPRESSOR/DNA COMPLEX + S-ADENOSYL-METHIONINE
{{The crystal structure of the met repressor-operator complex shows two dimeric repressor molecules bound to adjacent sites 8 base pairs apart on an 18-base-pair DNA fragment. Sequence specificity is achieved by insertion of double-stranded antiparallel protein beta-ribbons into the major groove of B-form DNA, with direct hydrogen-bonding between amino-acid side chains and the base pairs. The repressor also recognizes sequence-dependent distortion or flexibility of the operator phosphate backbone, conferring specificity even for inaccessible base pairs. }}
About this Structure
1cma is a 4 chain structure with sequence from Escherichia coli. Full crystallographic information is available from OCA.
Reference
- Somers WS, Phillips SE. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992 Oct 1;359(6394):387-93. PMID:1406951 doi:http://dx.doi.org/10.1038/359387a0