Malaria Parasite Plasmodium falciparum Erythrocyte Binding Antigen 140
From Proteopedia
Contents |
Introduction
In 2010, malaria caused over 650,000 deaths.[1] While this disease is both preventable and curable, many of those that live in areas where the disease is endemic do not have access to such resources. Although there is a vaccine currently undergoing clinical trials, results are not expected until 2014.[1] The most recent vaccine research has focused on understanding a group of proteins in the erythrocyte-binding ligand (EBL) family. These proteins, which are found in the Plasmodium species, facilitate entry into erythrocytes during malarial infection by creating tight junctions between the host erythrocytes and parasite membranes. [2] There are four family members: erythrocyte-binding antigen 175 (PfEBA-175), erythrocyte-binding ligand 1 (PfEBL-1), erythrocyte-binding antigen 140 (PfEBA-140), and erythrocyte-binding antigen 181 (PfEBA-181).[2] PfEBA-140 binds glycophorin C on host erythrocytes,[2] which helps maintain erythrocyte shape and regulates membrane material properties.[3] Understanding the mechanism by which PfEBA-140 recognizes and engages glycophorin C on erythrocytes may lead to the future development of a new malaria vaccine.
General Structure
|
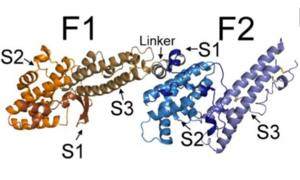
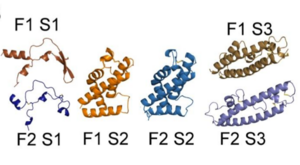
The EBL family members, including PfEBA-140, are made up of two regions, region II (RII) (shown to the right) and region VI.[2] Region II is responsible for receptor binding in all EBL family members.[2] RII is composed of , F1 (purple) and F2 (teal).[2] These two DBL domains are connected by a .[2] The DBL protein fold is unique to the Plasmodium species. Not only does it have the ability to recognize and bind many erythrocyte cell receptors, but it also mediates microvasculature adherence of infected erythrocytes by erythrocyte membrane protein 1 (PfEMP1).[2] Each DBL domain is composed of three subdomains, illustrated in the pictures below.
In the top image, the subdomains S1, S2, and S3 of each of the F1 and F2 domains, as well as the helical linker, are illustrated.[2] In the bottom image, structures of the individual subdomains are illustrated.[2]
The colors are the same in both images: F1 subdomain 1 is shown in bronze, subdomain 2 in orange, subdomain 3 in dark orange; F2 subdomain 1 is shown in dark blue, subdomain 2 in blue, and subdomain 3 in light blue.
Binding of RII PfEBA-140
|
Requirement of Both DBL Domains
Since the DBL domains of RII are highly conserved, the ability of the individual DBL domains to bind erythrocytes was tested. To do this, constructs containing the full-length RII PfEBA-140 and each individual DBL domain were tested using a rosetting assay. Both brightfield microscopy and green fluorescence protein (GFP) were used to visualize erythrocyte binding. In the upper panel, a construct expressing only GFP was used as a control. The lower panel of Figure A illustrates the extensive erythrocyte binding of the full length RII construct. In Figure B, the lack of black dots in the brightfield microscopy and the smaller quantity of green fluorescence illustrates that F1 and F2 are unable to independently bind erythrocytes. This result suggests that both domains equally participate in engaging erythrocytes.[2]Basic Patch
References
- ↑ 1.0 1.1 http://www.who.int/mediacentre/factsheets/fs094/en/index.html
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 Lin DH, Malpede BM, Batchelor JD, Tolia NH. Crystal and Solution Structures of Plasmodium falciparum Erythrocyte-binding Antigen 140 Reveal Determinants of Receptor Specificity during Erythrocyte Invasion. J Biol Chem. 2012 Oct 26;287(44):36830-6. doi: 10.1074/jbc.M112.409276. Epub 2012, Sep 18. PMID:22989878 doi:10.1074/jbc.M112.409276
- ↑ http://en.wikipedia.org/wiki/Glycophorin_C