This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 653
From Proteopedia
| This Sandbox is Reserved from 30/08/2012, through 01/02/2013 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 636 through Sandbox Reserved 685. | ||||||
To get started:
More help: Help:Editing For more help, look at this link: http://proteopedia.org/w/Help:Getting_Started_in_Proteopedia
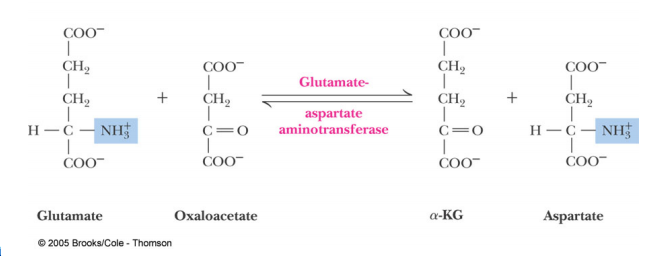
Glutamate-Aspartate Aminotransferase
Introduction: Aspartate Aminotransferase(AST)also known as serum glutamic oxaloacetic transaminase (SGOT)is an enzyme that functions within the Amino Acid Biosynthesis pathway to interconvert Glutamate and Aspartate. Aminotransferases work to transform amino acids using transamination reactions. This is a process which utilizes the exchange of an α-keto acid to alter an amino acid usually Glutamate and oxaloacetate to a second amino acid in this case Aspartate and α-ketoglutarate. The reaction is dependent on pyridoxial phosphate (PLP) a cofactor which switches between the Pyridoxial Phosphate(PLP) form and the Pyridoxamine Phosphate (PMP) form. The reaction is important in both amino acid synthesis and degradation. When looking at the formation of aspartate it is synthesized from oxaloacetate which is a key step of the citric acid cycle. Also glutamate can be degraded into ammonium ions through the oxidative deamination process. Aspartate Aminotransferae is generally found and has been studied in E.coli, pig heart cytosol, and chicken mitochondira. It is also present in other microorganisms such as Thermus thermophilus and Haloferax. However within these organisms it is commonly found in the muscles, tissues, heart,liver, and kidneys. This is why Aspartate Aminotransferase is so important in tests that monitor liver disease and damage to any of the above organs. [2]
Structure: The structure of Aspartate Aminotransferase has been determined through studies using X-ray crystallography. AST contains both α-helices packed on either side of a central sheet of β-strands in each of its domains. It is seen that AST has two identical subunits. Each subunit is approximately 300 residues long and each is composed of both a small and large domain. In addition a third domain is present which consists of the N-terminal residues 3-14; these residues link and stabilize the two subunits by forming a strand. Pyridoxial phosphate (PLP), the cofactor for AST binds to the large domain, interacting with the amino group of the Lys258 residue. It also interacts through hydrogen bonding with the Asp222 and Tyr 225 residues within the large domain. The small domain is responsible for the shift of the enzyme from an open conformation to closed conformation. This shift is based on the binding of the substrate. are located between the two domains near the interface. There are two independent active sites each contains two arginine residues which are responsible for the enzyme’s specificity for the substrate. The first is Arg 292 which forms complexes with carboxylate side chains and the second is ARg286 which interacts with the alpha carobxylate group of the substrate.[2]
Mechanism of Structure: Alpha ketoglutarate is an enzyme from Kreb’s Cycle, which exhibits several biochemical pathways. It reacts with a free amino acid that comes in the form of ketoacids. In addition, a cofactor Pyridoxal-5’-phosphate is added into the reaction in order to yield Glutamate and alpha ketoglutarate within the reaction of transamination by adding an amine to the alpha carbon carbonyl. This reaction involves another amino acid aspartate that uses another enzyme from the TCA cycle oxaloacetate, a reversible step enzyme that interconverts the appropriate amino acid while adding an amine to the actual precursor amino acid. Both of these reactions change the cofactor Pyridoxal-5’-phosphate to Pyridoxal-5’-amine which alters several conformational changes of the protein binding site. The actual cofactors P5P, PLP bind covalently and reversibly to an active-site in order to facilitate enzymatic reaction while the amino group is being added in to precursor used as a substrate. This helps catalyze the reaction by accepting the amino group from the amino acid then transferring it to the carbonyl compound of alpha keto glutarate and a keto acid to yield glutamate , then oxaloacetate and another form alpha keto acid to form an aspartate. There are other cofactors used in this reaction as intermediates through amination for yielding amino acids and regenerating the enzyme precursor from the Kreb’s cycle. [3][5]
Kirsch, Jack F. "Mechanism of Action of Aspartate Aminotransferase Proposed on the Basis of Its Spatial Structure." Journal Of Molecular Biology. Science Direct, 15 Apr. 1984. Web. 20 Nov. 2012. <http://www.sciencedirect.com/science/article/pii/0022283684903334>.[2] Rose, Bob. "Amino Acid Biosynthesis." Moodle, 15 Nov. 2011. Web. 20 Nov. 2012. <http://moodle.wolfware.ncsu.edu/file.php/30769/Day23_aa_biosynthesis_2012.pdf>.[3] Thompson, Gregory. "Aspartate Aminotransferase (AST)." Aspartate Aminotransferase. WebMD, 04 Nov. 2011. Web. 20 Nov. 2012. <http://www.webmd.com/digestive-disorders/aspartate-aminotransferase-ast>.[4] Aspartate Transaminase. Wikipedia, n.d. Web. 24 Nov. 2012. <http://en.wikipedia.org/wiki/Aspartate_transaminase>.[5] |