The human Bcr protein contains 1,271 amino acids and multiple domains, including the oligomerization domain (OLI), the serine/threonine kinase domain (S/TK), the domain homologous to the human Dbl and yeast Cdc24 proteins (DH) and the domain with GTPase-activating activity for Rac (RacGAP). The c-Abl protein contains 1,097 amino acids, which include the Src-homology domains 3/2 (SH3/SH2), tyrosine kinase domain (TK), nuclear translocalization signal (NTS), DNA binding domain (DB) and actin-binding motif (AB). Two domains are essential for transforming activity — OLI from Bcr and TK from Abl. Depending on the site of the breakpoint in the Bcr gene, the fusion protein can vary in size from 190 to 230 kDa. Each fusion protein contains the same portion of the c-Abl protein but differs in the length of the Bcr portion. [9]
Normal ABL has a tri-dimensional structure which is tightly preserved in a closed, inactive conformation order to prevent oncogenic activation. The maintenance of this inactive conformation is possible by:
1- The "latching" of the myristilated NH2-terminal sequence which is directly linked to a myristilation recognition sequence on the c-lobe of the SH1 kinase domain,
2- The close contact between SH3 and SH2 domain,
3- The interactions between SH3 domain and the C-lobe of the kinase domain. These interactions clamp the structure and prevent the kinase to switch to an active conformation, a process which requires the phosphorylation of Tyr 412 residue and the "unlatching" of the myristoyl group from the C-Lobe of the kinase domain. The attachment of proline-rich SH2 and SH3 ligands leads to the complete switch of the protein to an open, active conformation of the kinase. The NH2-terminal myristilation (autoregulatory role) is deleted during the t(9;22) translocation. [10]
Bcr-Abl tyrosine-kinase inhibitors
1st Generation TKIs: Gleevec
Imatinib mesylate (STI571 or Gleevec), discovered in 1992, is a Bcr-Abl tyrosine kinase inhibitor (Bcr-Abl TKI) and it's the most common first generation drug used for the CML treatment. Imatinib has a high affinity for ABL kinase.
Imatinib achieves BCR-ABL kinase inhibition by binding to the inactive, unphosphorylated conformation of the kinase (DFG-out)</scene> , thereby reducing the availability of the catalytically active phosphorylated conformation (DFG-in), necessary for ATP binding. This blocks signal transduction, ultimately resulting in inhibition of proliferation and loss of viability and proliferation.[11].
Imatinib is used for treating CML, gastrointestinal stromal tumours and other diseases. By 2011, Gleevec has been FDA approved to treat ten different cancers.
Pharmacokinetics: Imatinib is rapidly absorbed when given by mouth, and is highly bioavailable: 98% of an oral dose reaches the bloodstream. Metabolism of imatinib occurs in the liver and is mediated by several isozymes of the cytochrome P450 system. The main metabolite, N-demethylated piperazine derivative, is also active. The half-lives of imatinib and its main metabolite are 18 and 40 hours, respectively.
Adverse effects: The most common side effects include weight gain, reduced number of blood cells (neutropenia, thrombocytopenia, anemia), headache, edema, nausea, rash, and musculoskeletal pain.
Second generation TKIs: Nilotinib, Dasatinib
Nilotinib
Nilotinib in binding site
Nilotinib (AMN107) is a second-generation oral TKI. It was designed based on the crystal structure of imatinib to be highly active against a wide range of imatinib-resistant BCR-ABL mutants and is approved for the treatment of newly diagnosed or imatinib-resistant or -intolerant CML.
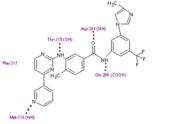
Nilotinib was designed to maintain binding to the inactive conformation of the ABL kinase domain, while incorporating alternative binding groups to the N-methylpiperazine moiety and preserving an amide pharmacophore to retain H-bond interactions with Glu286 and Asp381.
[12]
Substitution of the N-methylpiperazine moiety present in imatinib by a phenyl group bearing trifluoromethyl and imidazole substituents in the nilotinib structure greatly contributes to the potency of nilotinib by reducing the requirement for hydrogen bonding with nilotinib (four H-bond interactions compared with six H-bonds for imatinib).
Among patients with imatinib resistant CML, nilotinib has not been associated with the toxic effects commonly seen with imatinib treatment, such as fluid retention, edema, and weight gain [13]
Dasatinib
Dasatinib is also a second-generation TKI. Dasatinib binds kinases of the SCR family (0,2 nM<IC50<1,1nM). It binds both the active and inactiv forms of the Abl kinase with a higher affinity (300 times) than Imatinib. Dasatinib is active against most BCR-ABL mutants with the exception of T315I. [14] Dasatinib is well tollerated but adverse effects can include muscle and joint aches, fatigue, dermatologic complaints such as rash and acne, headaches, and diarrhea [15]
Third generation drugs
New mutations of Bcr-Abl tyrosine kinase are still being discovered and some old go unconquered. To overcome those mutations, new generation drugs are being developed to treat patients with CML.
Bosutinib (SKI-606)
On September 4, 2012, the U. S. Food and Drug Administration approved bosutinib tablets (Bosulif, Pfizer, Inc.) for the treatment of chronic, accelerated, or blast phase Philadelphia chromosome positive (Ph+) chronic myelogenous leukemia (CML) in adult patients with resistance or intolerance to prior therapy. [16]
is based on a quinoline scaffold and is structurally related to the AstraZeneca quinazoline template. [17]
Ponatinib (AP24534)
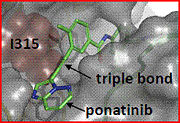
Ponatinib was identified using structure base drug design and focused synthetic libraries of trisubstituted purine analogs. The substance potently inhibits, on nanomolar scale, Src and Bcr-Abl kinases including many common imatinib resistant Bcr-Abl mutations, like T315I mutation. The key structural feature of the molecule is a carbon-carbon triple bond linkage that makes productive hydrophobic contact with the side chain of I315, allowing inhibition of the T315I mutant. The triple bond also acts as an inflexible connector that enforces correct positioning of the two binding segments of AP24534 into their established binding pockets. AP24534 maintains an extensive hydrogen-bonding network and occupies a region of the kinase that overlaps significantly with the imatinib binding site. [18]
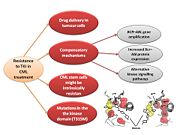
Resistance
Resistance to first generation TKI (Imatinib)
In the majority of cases, resistance is caused by reactivation of Bcr–Abl kinase activity. There are several mechanisms currently known that may result in Imatinib resistance [19]:
1. Plasma protein binding: Imatinib binds very strongly to the ∂-1-acid glycoprotein 1. Changes in ∂-1-acid glycoprotein may change the amount of binding of this drug, thus changing drug availability. This process can be also classified as a pharmacokinetic parameter.
2. Drug efflux: Imatinib is a substrate for the P-glycoprotein pump, which can result in decreased intracellular concentrations of Imatinib.
3. T315I mutation: In the presence of Imatinib, cells that generate mutations in BCR-ABL can overcome the ability of this drug to inhibit cell division. Mutations that alter the Imatinib binding site without affecting the adenosine triphosphate binding site or the active site of the kinase are very effective at inducing drug resistance. The gatekeeper in c-Abl kinase is a smaller threonine (Thr 315) that is not an effective stabilizer of the R-spine, but mutation in this residue is the most common mechanism implicated in secondary drug resistance. Usually, the gatekeeper is substituted by isoleucine or methionine and avoids Gleevec entrance to ATP-binding domain.
4. P-loop mutation: the structure of Bcr-Abl contains two flexible loops, the ATP-binding P-loop and the activation loop. Mutations in these loops destabilize arrangement of the loops such that the kinase domain cannot assume the inactive conformation required for Imatinib binding. There are clinical data indicating that Bcr-Abl mutations in the P-loop is 70-100 fold less sensitive to imatinib compared with native Bcr-Abl. [20]
5. Activation of different kinases: although the 9:22 translocation is necessary to initiate CML, BCR-ABL is only one of several kinases capable of maintaining the proliferation rate of the cell while inhibiting apoptosis.
6. Gene amplification: as the number of Philadelphia chromosomes increases, the number of BCR-ABL proteins expressed in the cell increases and the efficacy of Imatinib decreases.
7. Inadequate plasma levels of Imatinib: Subtherapeutic dosing is highly likely to result in the selection of a resistant clone. That’s why measurement of plasma levels is very important. To solve this problem second generation drugs (Nilotinib) were developed.
8. Pharmacokinetic parameters: The cytochrome P450 enzymes metabolize Imatinib by the CYP3A4 isoform. The variability in CYP3A4 activity among individuals is may contribute to some of the variation in imatinib levels between patients. In addition, other drugs taken by patients can alter CYP3A4 activity. Further polymorphism studies are mandated to understand the significance of cytochrome P450 enzymes on the plasma levels of Imatinib and response to therapy in CML patients.
Resistance to second generation of TKI (Nilotinib/Dasatinib)
Nilotinib and Desatinib are ineffectiveness against the T315I mutant. It is important to underline that all mutations except T315I were effectively suppressed by increasing Nilotinib concentration. Although Dasatinib is much more potent than Imatinib it is possible, like with Nilotinib, that its specific mode of binding to Abl may lead to new vulnerable sites that could confer new kinds of drug resistance. Mutations have been found on Phe317 so that is a potential vulnerable site for this drug [21]
Resistance to drugleads
T315 mutation is resistant to Bosutinib. In contrast to imatinib, nilotinib and dasatinib, bosutinib is not an efficient substrate for multidrug resistance (MDR) transporters that promotes efflux of foreign molecules from cells. Bosutinib even inhibits these transporter proteins in higher concentrations. [22]
New strategies in controlling drug resistance
Dealing with the rise of these resistant clones has presented an important challenge to health care providers. Thus, new therapeutic strategies have been developed (New strategies in controlling drug resistance:
• Bone marrow transplantation: Imatinib and Dasatinib are tremendously effective at blocking disease progression; however, they are generally not thought to be curative. Currently, the only known curative therapy is bone marrow transplant. However, this is only a potential option for younger patients in good performance status.
• Stopping Imatinib after cytgenetic remission: in patients who achieve a complete molecular response (CMR), Bcr-Abl transcripts are no longer detectable by PCR.
• Allogeneic HCT (AlloHCT): use for those who are intolerant to TKIs or have mutations such as the T315I mutation, which can produce significant resistance to all of the clinically available TKIs. In addition, it seems that in high-risk or advanced-phase patients, a more aggressive approach would be to combine second-generation TKIs initially, followed by AlloHCT. [23] .
• Autologous stem cell transplantation (auto-SCT): could also eliminate a Ph+ clone bearing a BCR-ABL kinase domain mutation, thus could be a promising therapeutic option for imatinib resistance.
• ATP non-competitive ABL TKI: useful for patients with the T315I mutation. Some examples are ON012380, the aurora kinase inhibitor MK-0457, and the p38 MAP kinase inhibitor BIRB796 [24] .
Current situation
Now a days, several estudies are being developed. For further information about the pipeline, check clinical trials.gov
Authors
Julio César Corral; Almudena Chaves Pérez; Cristina Murga Montesinos.
References
- ↑ Yuan ZM, Huang Y, Ishiko T, Kharbanda S, Weichselbaum R, Kufe D. Regulation of DNA damage-induced apoptosis by the c-Abl tyrosine kinase. Proc Natl Acad Sci U S A. 1997 Feb 18;94(4):1437-40. PMID:9037071
- ↑ Wang JY. Regulation of cell death by the Abl tyrosine kinase. Oncogene. 2000 Nov 20;19(49):5643-50. PMID:11114745 doi:10.1038/sj.onc.1203878
- ↑ Crystal Structures of the Kinase Domain of c-Abl in Complex with the Small Molecule Inhibitors PD173955 and STI571
- ↑ Schindler T, Bornmann W, Pellicena P, et al. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938 – 42.
- ↑ [Ramakrishnan, C. et al. (2002) A conformational analysis of Walker motif A [GXXXXGKT (S)] in nucleotide-binding and other proteins. Protein Eng. 15, 783–798]
- ↑ [Johnson, D.A. et al. (2001) Dynamics of cAMP-dependent protein kinase. Chem. Rev. 101, 2243–2270].
- ↑ KIT kinase mutants show unique mechanisms of drug resistance to imatinib and sunitinib in gastrointestinal stromal tumor patients
- ↑ Leukemia research 34 (10): 1255–1268. doi:10.1016/j.leukres.2010.04.016. PMID 2053738
- ↑ Zhao X, Ghaffari S, Lodish H, Malashkevich VN, Kim PS. Structure of the Bcr-Abl oncoprotein oligomerization domain. Nat Struct Biol. 2002 Feb;9(2):117-20. PMID:11780146 doi:10.1038/nsb747
- ↑ http://atlasgeneticsoncology.org/Genes/ABL.html
- ↑ Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol. 2006;2:358 – 64.
- ↑ Weisberg E, Manley PW, Breitenstein W, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129 – 41.
- ↑ Kantarjian H, Giles F, Wunderle L, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosomepositive ALL. N Engl J Med. 2006;354:2542–51.
- ↑ Shah NP, Tran C, Lee FY, et al. (2004) Overriding imatinib resistance with a novel ABL kinase inhibitor. Science 305:399–401.
- ↑ Results of Dasatinib Therapy in Patients With Early Chronic-Phase Chronic Myeloid Leukemia Jorge E. Cortes, Dan Jones, Susan O'Brien, Elias Jabbour, Farhad Ravandi, Charles Koller, Gautam Borthakur, Brenda Walker, Weiqiang Zhao, Jianqin Shan and Hagop Kantarjian
- ↑ http://www.fda.gov/Drugs/InformationOnDrugs/ApprovedDrugs/ucm318203.htm
- ↑ Manley, P.; Cowan-Jacob, S.; Mestan, J. (2005). "Advances in the structural biology, design and clinical development of Bcr-Abl kinase inhibitors for the treatment of chronic myeloid leukaemia". Biochimica et Biophysica Acta 1754 (1–2): 3–13.
- ↑ O'Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, Adrian LT, Zhou T, Huang WS, Xu Q, Metcalf CA 3rd, Tyner JW, Loriaux MM, Corbin AS, Wardwell S, Ning Y, Keats JA, Wang Y, Sundaramoorthi R, Thomas M, Zhou D, Snodgrass J, Commodore L, Sawyer TK, Dalgarno DC, Deininger MW, Druker BJ, Clackson T. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009 Nov 6;16(5):401-12. PMID:19878872 doi:10.1016/j.ccr.2009.09.028
- ↑ (New strategies in controlling drug resistance)
- ↑ An X, Tiwari AK, Sun Y, Ding PR, Ashby CR Jr, Chen ZS. BCR-ABL tyrosine kinase inhibitors in the treatment of Philadelphia chromosome positive chronic myeloid leukemia: a review. Leuk Res. 2010 Oct;34(10):1255-68. PMID:20537386 doi:10.1016/j.leukres.2010.04.016
- ↑ (Olivieri, A.; Manzione, L. (2007). "Dasatinib: a new step in molecular target therapy". Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 18 Suppl 6: vi42–vi46)
- ↑ Boschelli, F.; Arndt, K.; Gambacorti-Passerini, C. (2010). "Bosutinib: a review of preclinical studies in chronic myelogenous leukaemia". European journal of cancer (Oxford, England : 1990) 46 (10): 1781–1789.doi:10.1016/j.ejca.2010.02.032. PMID 20399641. edit)
- ↑ [J. Cortes, D.W. Kim, E. Raffoux, G. Martinelli, E. Ritchie, L. Roy et al. Efficacy and safety of dasatinib in imatinib-resistant or -intolerant patients with chronic myeloid leukemia in blast phase Leukemia, 22 (2008), pp. 2176–2183].
- ↑ (BCR-ABL tyrosine kinase inhibitors in the treatment of Philadelphia chromosome positive chronic myeloid leukemia: A review; Xin An, Amit K. Tiwari, Yibo Sua, Pei-Rong Ding, Charles R. Ashby Jr., Zhe-Sheng Chen)