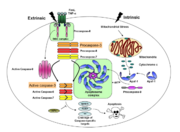
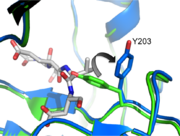
Exosite and Allosteric Site
Since all caspases have very similar active site chemistry, other regions in the protein that can confer either activation or inhibition to the enzyme needs to be explored. For example, exosites in caspase-7 have been identified and were observed to improve activity (Boucher, Blais et al. 2012). Caspase-7 also has an inhibitory allosteric site that could bind with the small molecule FICA, resulting in a zymogen-like conformation (Hardy, Lam et al. 2004) and abolishing activity.
Although there are still no evident exosites found in caspase-3, some allosteric sites, most of which are located on the dimer interface, have been interrogated by mutagenesis and have been shown to modulate the activity of caspase-3 or even procaspase-3. Although only procaspase-3 was detected with little activity because the orientation of ILA (prematured L2 loop) and ILB loop cannot form an active site pocket, it is sill quite interesting to discover how these mutations are able to rescue activity of the zymogen form (Bose, Pop et al. 2003).
One interesting mutation, V266E, improves caspase-3 activity dramatically. Even in the uncleavable procaspase-3 (D5A, D26A, D175A), V266E mutant zymogen is still pseudo-activated, showing a 60-fold increase in activity. Intriguingly, V266E does not undergo many conformational changes around the active site in its cleaved form. Based on the crystal structure of this mutant, the L2’ loop is partially disordered at 185’-180’. Moreover, this active procaspase-3 variant cannot be inhibited by endogenous XIAP like normal cleaved caspase-3. So it provides an option for apoptosis stimuli with intrinsic efficiency.
It was found recently that changing other critical residues (V266H, Y197C, E124A) on the dimer interface might play an important role on inhibition of caspase-3 by altering hydrogen bond patterns or through a relay of subtle conformational changes that spreads across the whole dimer.
Post translational Modification
Caspase-3 has been reported to be phosphorylated at Ser-150 by p38-MAPK and directly inhibits its activity. Conversely, phosphorylation of caspase-3 by protein kinase C zeta (PKC-ζ) was shown to promote autocleavage and activation.
Aside from phosphorylation, caspase-3 is also known to be ubiquitinylated and nitrosylated, however the underlying mechanismsand effect of these modifications are still unclear.
Natural Inhibitors
X-linked inhibitor of apoptosis proteins (XIAP) contains the second baculovirus IAP repeat domain (BIR2) targeting caspase-3 and caspase-7. The domain sits directly in the active site of caspase-3 and completely inhibits the protein. The region binding the active site runs in the opposite direction of normal caspase-3 substrates, thus occupying P1 through P4 but avoiding cleavage by the protease. This unique method of inhibition is a critical regulatory mechanism used in cells to control apoptotic caspase activity.
References
Bose, K., C. Pop, et al. (2003). "An uncleavable procaspase-3 mutant has a lower catalytic efficiency but an active site similar to that of mature caspase-3." Biochemistry 42(42): 12298-12310.
Boucher, D., V. Blais, et al. (2012). "Caspase-7 uses an exosite to promote poly(ADP ribose) polymerase 1 proteolysis." Proc Natl Acad Sci U S A 109(15): 5669-5674.
Hardy, J. A., J. Lam, et al. (2004). "Discovery of an allosteric site in the caspases." Proc Natl Acad Sci U S A 101(34): 12461-12466.