This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
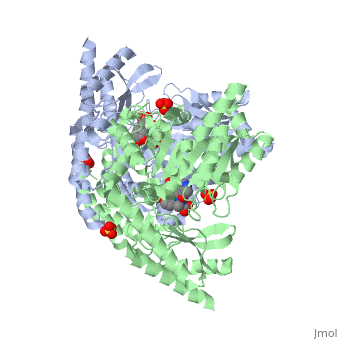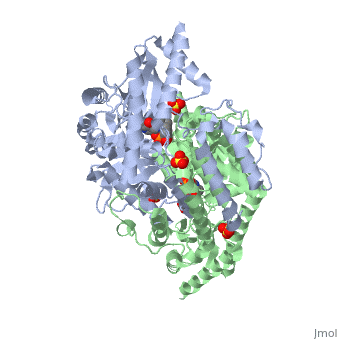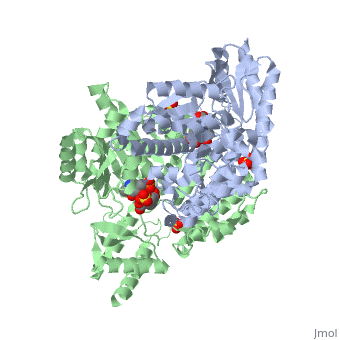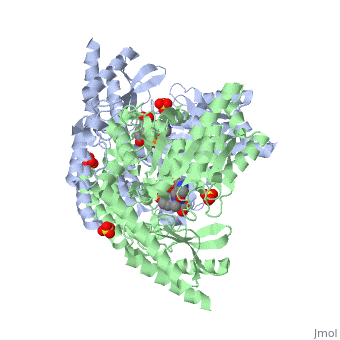
1js3
From Proteopedia
| |||||||||
| 1js3, resolution 2.25Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Activity: | Aromatic-L-amino-acid decarboxylase, with EC number 4.1.1.28 | ||||||||
| Related: | 1js6 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Contents |
Crystal structure of dopa decarboxylase in complex with the inhibitor carbidopa
Template:ABSTRACT PUBMED 11685243
About this Structure
1js3 is a 2 chain structure with sequence from Sus scrofa. Full crystallographic information is available from OCA.
See Also
Reference
- Burkhard P, Dominici P, Borri-Voltattorni C, Jansonius JN, Malashkevich VN. Structural insight into Parkinson's disease treatment from drug-inhibited DOPA decarboxylase. Nat Struct Biol. 2001 Nov;8(11):963-7. PMID:11685243 doi:http://dx.doi.org/10.1038/nsb1101-963