We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Michael Roberts/BIOL115 Myo
From Proteopedia
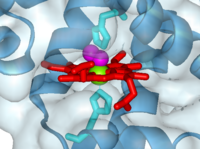
Myoglobin with oxygen bound to heme (1a6m)
The heme group and oxygen binding in myoglobin.
Myoglobin is a protein whose function is to store oxygen in muscle tissues. Like heamoglobin, it is red in colour, and it is myoglobin that gives muscle its strong red colour.
Myoglobin was the first globular protein for which the 3-dimensional structure was solved, back in the late 1950s. It gives its name to the 'globin fold', a common alpha domain motif. An alpha domain is a structural region composed entirley of alpha-helix.
Click on the 'green links' in the text in the scrollable section below to examine this molecule in more detail.
| |||||||||||
