We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
From Proteopedia
proteopedia linkproteopedia link
| This Sandbox is Reserved from 30/01/2013, through 30/12/2013 for use in the course "Biochemistry II" taught by Hannah Tims at the Messiah College. This reservation includes Sandbox Reserved 686 through Sandbox Reserved 700.
|
To get started:
- Click the edit this page tab at the top. Save the page after each step, then edit it again.
- Click the 3D button (when editing, above the wikitext box) to insert Jmol.
- show the Scene authoring tools, create a molecular scene, and save it. Copy the green link into the page.
- Add a description of your scene. Use the buttons above the wikitext box for bold, italics, links, headlines, etc.
More help: Help:Editing
|
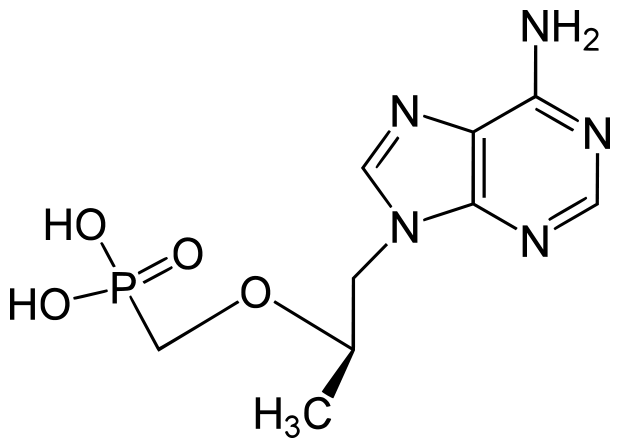
Complera: Emtricitabine/rilpivirine/tenofovir
Reverse Transcriptase:
Upon infection, HIV binds to the CD4 receptor in a helper T cell or other CD4-presenting lymphocyte. The viral envelope is then fused with the cellular membrane allowing intracellular release of viral particles. The utilization of several enzymes helps to solidify the virus’s hold on the cell. HIV reverse transcriptase (RT) works to convert the viral single-stranded RNA (ssRNA) into DNA. HIV integrase then helps to incorporate the newly reverse transcribed DNA into the cellular genome. (Voet et al. 2008)
The structure of HIV-1 reverse transcriptase is a dimeric protein with a 66-kD polymerase (p66) domain and a 51-kD RNase H domain (p51). The enzyme has a conformation and possesses one active site and three catalytic functions: RNA-dependent DNA polymerization, RNA hydrolysis, and DNA replication. The p66 domain is an RNA-dependent DNA polymerase which binds viral RNA as well as DNA in its single palm-like active site. This active site is lined with positive charges (blue) which interact with the negatively charged phosphodiester backbone of the . This diversity of function is brought about by the flexible nature of the hand-like p66 subunit. The p51 subunit is the RNase H domain which degrades the viral RNA after a daughter DNA strand has been synthesized. A structural overview helps us understand the basis of the high mutability of HIV. RNA, which forms the basis of the viral genetic material, is less stable than DNA and is more subject to modification or damage. In addition the reverse transcriptase polymerase domain lacks proofreading exonuclease activity. This lowers the fidelity of the reverse transcription. (Voet et al. 2008, Abbondanzieri et al. 2008)
RPV in the
In red are the binding pocket residues p66 (Leu-100, Lys-101, Lys-103, Val-106, Thr-107, Val-108, Val-179, Tyr-181, Tyr-188, Val-189, Gly-190, Phe-227, Trp-229, Leu- 234, and Tyr-318) and p51(Glu-138). Singh et al.
Image:Emtricitabine3.png

