User:Samuel Carlson/Sandbox 1
From Proteopedia
Contents |
Bromodomain adjacent to zinc finger domain protein 2A (BAZ2a)
Overview
| |||||||||
| Human BAZ2b bromodomain structure, BAZ2a bromodomain is assumed similar 3g0l | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Gene: | BAZ2B (Homo sapiens) | ||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
BAZ2A is a transcription factor which acts as a regulator of chromatin remodeling. It is part of the nucleolar remodeling complex (NoRC).
This structure consists of a methyl-CpG binding domain (MBD) near the N-terminus, a DNA-binding homeobox and different transcription factor (DDT) domain, and a plant homeodomain (PHD) zinc finger next to a bromodomain on the N-terminus.
Role in the Nucleolar Remodeling Complex (NoRC)
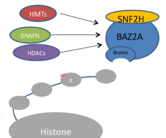
BAZ2A interacts with sucrose-nonfermenting protein 2 homolog (SNF2H) to form the NoRC, which recruits histone methyltransferases, histone deacetylases[1], and DNA methyltransferases. The NoRC silences transcription by modifying DNA compaction, as well as by interfering with ribosomal RNA (rRNA) transcription. BAZ2a is thought to direct transcriptional repression by binding to histones labeled with posttranslational modifications (PTMs) such as acetylation on lysine residues.[2]
Evolution and related structures
The bromodomain is a highly conserved structure within BAZ2a. It can be found in many large proteins associated with helicase activity, chromatin remodeling, methyl- or acetyltransferase activity, or transcriptional control. [3] The structures of individual domains (PHD fingers, Bromodomains, MBD domains, and DDT domains) similar to those found in BAZ2a have been solved. Additional domain structures can be found at pages linked below, for PHD finger and Bromodomain categories on Proteopedia.;
BAZ2a Function
Histone Recognition
The PHD finger domain of BAZ2a may also bind histones, as its structure and sequence are similar to those of other PHD fingers, which have been shown to bind to methylated lysine residues on H3. [6][7]
The PHD finger and Bromodomain may act together to recognize posttranslationally modified histones, as is the case in the human Bromodomain PHD transcription factor BPTF.[8]
DNA and RNA binding
Because of a methyl-CPG binding domain and two AT hooks, BAZ2a binds to DNA with high affinity, with no specificity for methylation.[9]
Role in Chromatin Remodeling
As a critical member of the NoRC, BAZ2a in conjunction with SNF2H is responsible for modifying the compaction of chromatin, through The NoRC is also responsible for ribosomal DNA (rDNA) transcription silencing, by recruiting histone deacetylase (HDAC) and DNA methyltransferase (DNMT) to the rDNA promoter, effectively creating a region with the characteristics of heterochromatin and silencing rDNA transcription. [10]
Medical Implications
Recent studies have shown that BAZ2a depletion causes suppression of chromatin remodeling and increases rRNA transcription. [11] Loss of BAZ2a functionality can therefore lead to genetic instability, phenotypic transformation, and cell proliferation beyond confluence. [12] Other bromodomain families have been identified as potent drug targets for inhibitors[13], specifically at their acetyl-lysine binding pockets, due to certain amino acid signatures. [14] However, the bromodomain of BAZ2a and other bromodomains with more evolutionarily similar sequences have not been investigated yet.
References
- ↑ Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006 May 5;22(3):351-61. PMID:16678107 doi:10.1016/j.molcel.2006.03.028
- ↑ Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012 Mar 30;149(1):214-31. PMID:22464331 doi:10.1016/j.cell.2012.02.013
- ↑ Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Muller S, Pawson T, Gingras AC, Arrowsmith CH, Knapp S. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012 Mar 30;149(1):214-31. PMID:22464331 doi:10.1016/j.cell.2012.02.013
- ↑ Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999 Jun 3;399(6735):491-6. PMID:10365964 doi:10.1038/20974
- ↑ Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999 Jun 3;399(6735):491-6. PMID:10365964 doi:10.1038/20974
- ↑ Shi X, Kachirskaia I, Walter KL, Kuo JH, Lake A, Davrazou F, Chan SM, Martin DG, Fingerman IM, Briggs SD, Howe L, Utz PJ, Kutateladze TG, Lugovskoy AA, Bedford MT, Gozani O. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J Biol Chem. 2007 Jan 26;282(4):2450-5. Epub 2006 Dec 1. PMID:17142463 doi:10.1074/jbc.C600286200
- ↑ Britton AM, Nichols JD, Draper PR. The comparative bioavailability of cimetidine-alginate treatments. J Pharm Pharmacol. 1991 Feb;43(2):122-3. PMID:1672897
- ↑ Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, Allis CD. Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions. Cell. 2011 May 27;145(5):692-706. Epub 2011 May 19. PMID:21596426 doi:10.1016/j.cell.2011.03.053
- ↑ Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I. NoRC--a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001 Sep 3;20(17):4892-900. PMID:11532953 doi:10.1093/emboj/20.17.4892
- ↑ Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet. 2002 Nov;32(3):393-6. Epub 2002 Oct 7. PMID:12368916 doi:10.1038/ng1010
- ↑ Santoro R, Lienemann P, Fussenegger M. Epigenetic engineering of ribosomal RNA genes enhances protein production. PLoS One. 2009 Aug 14;4(8):e6653. doi: 10.1371/journal.pone.0006653. PMID:19680546 doi:10.1371/journal.pone.0006653
- ↑ Guetg C, Lienemann P, Sirri V, Grummt I, Hernandez-Verdun D, Hottiger MO, Fussenegger M, Santoro R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010 Jul 7;29(13):2135-46. doi: 10.1038/emboj.2010.17. Epub 2010 Feb 18. PMID:20168299 doi:10.1038/emboj.2010.17
- ↑ Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, Philpott M, Munro S, McKeown MR, Wang Y, Christie AL, West N, Cameron MJ, Schwartz B, Heightman TD, La Thangue N, French CA, Wiest O, Kung AL, Knapp S, Bradner JE. Selective inhibition of BET bromodomains. Nature. 2010 Sep 24. PMID:20871596 doi:10.1038/nature09504
- ↑ Vidler LR, Brown N, Knapp S, Hoelder S. Druggability analysis and structural classification of bromodomain acetyl-lysine binding sites. J Med Chem. 2012 Sep 13;55(17):7346-59. doi: 10.1021/jm300346w. Epub 2012 Jul 12. PMID:22788793 doi:10.1021/jm300346w