BamHI
From Proteopedia
Overview
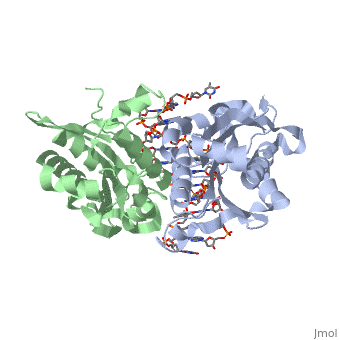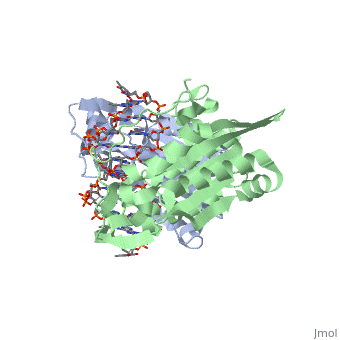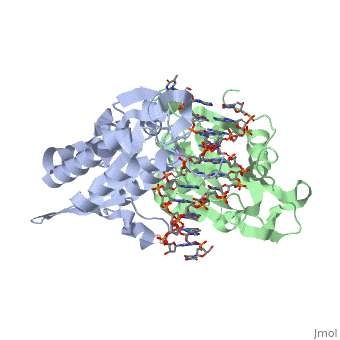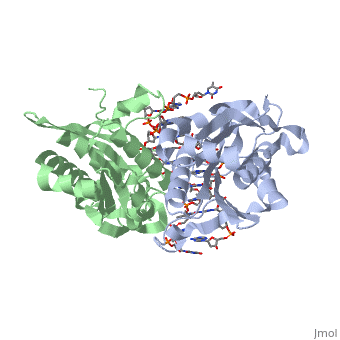
is a type II restriction enzyme derived from Bacillus amyloliquefaciens. Like all Type II restriction endonucleases, it is a and the recognition site is palindromic and 6 bases in length. It recognizes the DNA sequence of G’GATCC and leaves an overhang of GATC which is compatible with many other enzymes.[1] The BamHI-DNA complex is a sequence-specific endonucleases-DNA complex. In the complex all occuring in the major groove of the recognition site takes part in direct or water-mediated hydrogen bonds with the protein, namely on oxygens and nitrogens in the DNA within 3.5 angstroms of the protein. No other DNA sequence could support this degree of complementarity with BamHI.[2] There are five crystal structures of BamHI in the Protein Data Bank. These include BamHI bound to a non-specific DNA, BamHI complex with DNA and calcium ions (pre-reactive complex), BamHI complex with DNA and manganese ions (post-reactive complex), BamHI complex with DNA, and BamHI phased at 1.95 angstroms resolution by MAD analysis.[3] See also Endonuclease.
| |||||||||||
3D structures of BamHI
Updated on 10-October-2013
1bhm, 3bam, 2bam, 1esg – BAM + DNA – Bacillus amyloliquefaciens
References
- ↑ 1.0 1.1 Viadiu H, Aggarwal AK, Structure of BamHI bound to nonspecific DNA: a model for DNA sliding., Mol Cell. 2000 May;5(5):889-95. Print.
- ↑ Voet, Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry Life at the Molecular Level. New York: John Wiley & Sons, 2008. p. 578-579. Print.
- ↑ 3.0 3.1 PDBe., European Molecular Biology Laboratory. 2011. Webpage.
- ↑ Newman, Matthew, Structure of BamHI Endonuclease Bound to DNA: Partial Folding and Unfolding on DNA Binding., Science. 1995 Aug;4(269):656-63. Print.
Proteopedia Page Contributors and Editors (what is this?)
Shane Michael Evans, Michal Harel, Alexander Berchansky, Ann Taylor