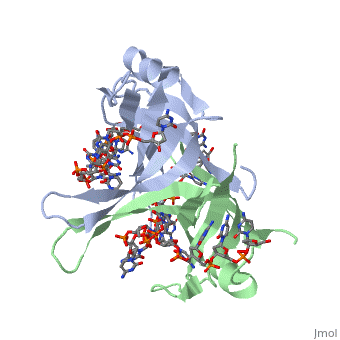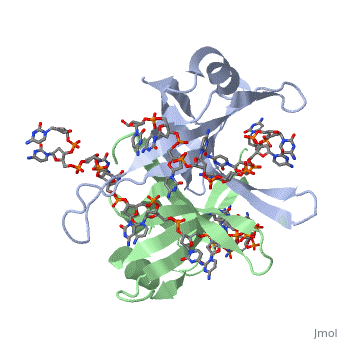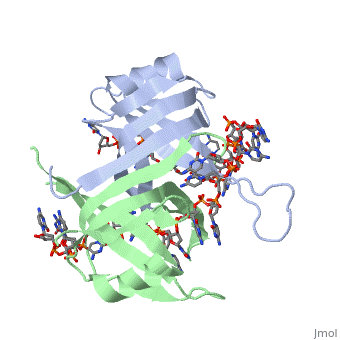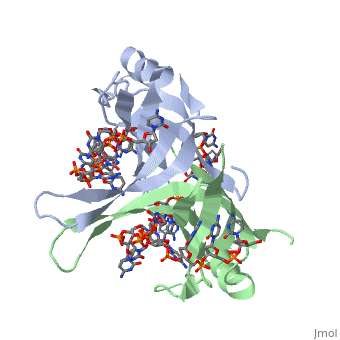
Binding Interactions in the Active Site
Phe-60 is an important DNA binding site. It has been shown to be the site for cross-linking.
Tryptophan and Lysine residues are important in binding as well. Treatments resulting in
modification of arginine, cysteine, or tyrosine residues had no effect on binding of SSB to
DNA, whereas modification of either lysine residues (with acetic anhydride) or tryptophan
residues (with N-bromosuccinimide) led to complete loss of binding activity ( Meyer, 348).
The two tryptophan residues involved in DNA binding are Trp40 and Trp54, which was
determined by mutagenesis. One more binding site was determined by site-specific mutagenesis.
When His55 is substituted with Leu it decreases binding affinity. All of these residues
are found in a hydrophobic region, which is suitable for nucleotide base interactions.
SSB-Protein Interactions
It is believed that Gly15 may play an important role in binding the RecA protein. Mutations in Gly15 have
extreme effects on recombinational repair. SSB has also been thought to bind with exonuclease I, DNA polymerase II,
and a protein n, which is used to help synthesize RNA primers for the lagging strand.