Sandbox Reserved 761
From Proteopedia
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
|
Contents |
Glutamate Dehydrogenase
Glutamate Dehydrogenase (GDH) is a homohexameric enzyme required for urea synthesis, that catalyses the reversible oxidative deamination of L-glutamate to α-ketoglutarate, and vice versa using NAD+ and/or NADP+ as coenzymes. Located in the mitochondria, GDH is an important branch-point enzyme between carbon and nitrogen metabolism. GDH catalyzes the reversible NAD (P)+-linked oxidative deamination of L-glutamate into alpha ketoglutarate and ammonia in two steps. The first step involves a Schiff base intermediate being formed between ammonia and alpha ketoglutarate. This Schiff base intermediate is crucial because it establishes the alpha carbon atom in glutamate’s stereochemistry. The second step involves the Schiff base intermediate being protonated, which is done by the transfer of a hydride ion from NADPH resulting in L-glutamate. GDH is unique because it is able to utilize both NAD+ and NADP+ [1]. NADP+ is utilized in the forward reaction of alpha ketogluterate and free ammonia, which are converted to L-glutamate via a hydride transfer from NADPH to glutamate (15). NAD+ is utilized in the reverse reaction, which involves L-glutamate being converted to alpha ketoglutarate and free ammonia via an oxidative deamination reaction [2]. The extensive production of ammonia by peripheral tissue or glutamate dehydrogenase is not allowed because of the highly toxic effects of circulating ammonia in cells. As a result, the ammonia produced in the reverse reaction of GDH is excreted as NH4+ in the urine, by first going through the urea cycle.
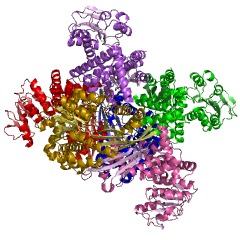
Glutamate Dehydrogenase Structure
GDH is a homohexamer of 505 residues with a molecular weight of 56 KDa (18). The structure of GDH is essentially two trimers of six identical subunits containing three distinct domains; the Glutamate (Glu) binding domain at the N terminus, the NAD binding doman, and the antenna domain, separated by a large active site cleft (9). The overall structure of GDH is composed of eighteen alpha helices and thirteen beta sheets, which are both parallel and anti-parallel and flanked by a layer of alpha helices (18).
The N-terminal glutamate binding domain,composed of mainly beta sheets,NAD-BD sits on the top of Glu-BD. NAD-BD and Glu-BD form the catalytic cleft. During substrate binding, the NAD-BD moves significantly. This movement has two components, rotating along the long axis of a helix at the back of the NAD-BD, called "the pivot helix", and twisting about the antenna in a clockwise fashion. A comparison of the open and closed conformations of GLUD1 reveals changes in the small helix of the descending strand of the antenna, which seems to recoil as the catalytic cleft opens.[1] Closure of one subunit is associated with distortion of the small helix of the descending strand that is pushed into the antenna of the adjacent subunit. R496 is located on this small helix (see Mutations).
The core structure of the hexamer is a stacked dimer of trimers. Glu-BDs of the monomers are mainly responsible in the build up of the core. The relative position of the monomers is such that the rotation about the pivot helix in each monomer is not restricted. The antennae from three subunits within the trimers wrap around each other and undergo conformational changes as the catalytic cleft opens and closes. The antenna serves as an intersubunit communication conduit during negative cooperativity and allosteric regulation.
NAD+ binding domains are located on top of the N and C-domains. These NAD+ binding domains rotate down upon the substrate and coenzyme to initiate catalysis. The NAD+ binding domain contains a forty eight-residue “antenna” that extends from the top of the NAD+ binding domain. This antenna undergoes conformational changes as the cleft of the active site opens and closes (2). Both of the domains are positioned differently in GDH. When GDH is not bound by glutamate its cleft is open, however when GDH is bound by glutamate it is closed. This position difference between the two domains allows the cleft to be closed, which brings the C4 of the nicotinamide ring and the alpha carbon of the glutamate substrate into the appropriate orientation for a hydride transfer to occur. Residues 200-206, 375-379, and 421-423 are critical for the control of the hinges that open or close the cleft between the two domains (1). The residues that form this hinge, which allow the cleft to open or close are both near and far from the active site. Gly 182, 183, Leu 185, and Gly 186 are near the active site, whereas Gly 361 is far from the active site. These five residues form two long loops connecting the two domains (9).
Glutamate Dehydrogenase Mechanism
The free energy change for the conversion of glutamate to alpha ketoglutarate is 3.7 kcal/mol (4). Although this ATP-consuming pathway is energetically unfavourable, this could be related to the continuation of re-dox equilibrium to re-oxidise the excess of NADH produced during glycolysis.