Sandbox Reserved 776
From Proteopedia
|
| This Sandbox is Reserved from Sep 25, 2013, through Mar 31, 2014 for use in the course "BCH455/555 Proteins and Molecular Mechanisms" taught by Michael B. Goshe at the North Carolina State University. This reservation includes Sandbox Reserved 299, Sandbox Reserved 300 and Sandbox Reserved 760 through Sandbox Reserved 779. |
To get started:
More help: Help:Editing |
Contents |
Dehaloperoxidase
Dehaloperoxidase (DHP) from the marine annellid Amphitrite ornata is a dimeric enzyme weighing ~16 kDa with a heme prosthetic group and belongs to the globin family. DHP is consistent with the 'globin fold', having eight alpha helices; globins are reversible oxygen binding/transport proteins (). Although belonging to the globin superfamily, DHP has a biologically relevant peroxidase activity, the first known globin-peroxidase. It has previously been shown to dehalogenate trihalophenols to their corresponding dihaloquinones. Peroxidases catalyze the reaction of various substrates utilizing hydrogen peroxide as the electron acceptor and [O] insertion to the product originates from water. Reactivity is initiated by use of H2O2 as co-substrate. 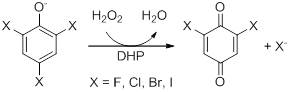
There are two known isoenzymes of DHP, A and B. DHP B has been shown to be more reactive than DHP A, although DHP A has been more extensively studied in the past. The isoenzymes differ at only five residues; I9L, R32K, Y34N, R81S and S91G. (1)(2)
Structure
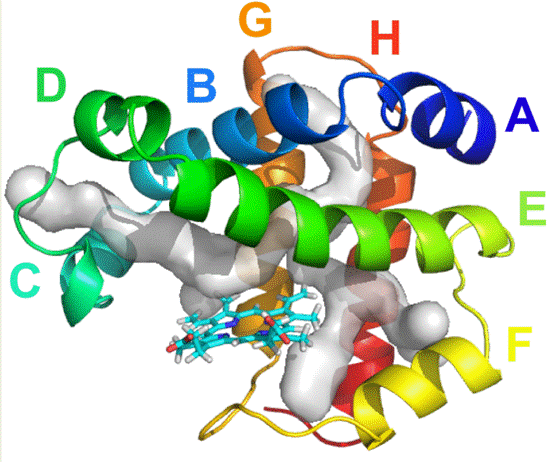
DHP is an enzyme with 127 residues and primarily a dimer in solution which has only been crystallized in its dimeric form. Although DHP belongs to the globin superfamily, with 8 alpha helices () and the final two helices form an anti-parallel helix-turn-helix motif, it is rich in aspartic and glutamic acids similar to class III peroxidases (2)(3).
Photo credit to group member Jing Zhao.
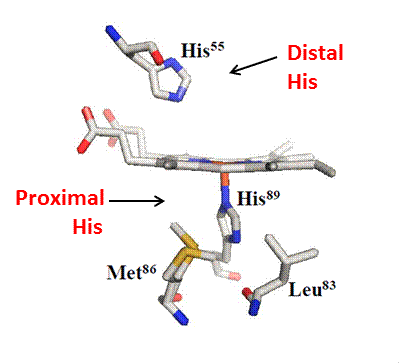
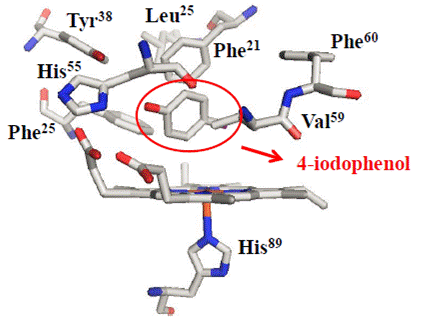
Important for the catalytic activity, DHP has proximal and distal histidines, H89 and H55 respectively. H89 is responsible for the only covalent linkage between heme iron and peptide chain in the fifth coordination site, while H55 is responsible for the stabilization and hydrogen bonding with axial ligands such as water, peroxide and cyanide. . On the proximal side, there is Met86 which creates a strong hydrogen bond to H89 and is apart of the backbone carbonyl-His-Fe catalytic triad which generates a neutral histidine like other globins; however, generates an enhanced peroxidase activity. . This interaction aids to the cleavage of the peroxide O-O bond to form reactive intermediates. In the distal pocket there are several hydrophobic residues; Phe21, Phe24, Phe35, Phe52 and Phe60 which assist the binding of organic molecules in the heme site. (2)(3)
Work to solve the structure of DHP mainly includes X-ray crystallography, however NMR studies have been done to see structure perdibation with substrates.
X-ray crystallography has been accomplished to determine wild type DHP apart from several mutants, as well as conformations of O2 bound heme (oxyferrous), the cyanide ligand complex and inhibitor/substrate bound protein. Crystallography was also able to show allosteric changes at the dimer interface upon addition of phenolic substrates. "1 H NMR experiments on the low-spin metcyano DHP showed different modes of binding between the mono-, di-, and trihalophenols as substrates. 13 C and 15 N labeled DHP showed that trihalogenated substrates altered the distal histidine and amide protons near tryptophan 120" (4). These experiments were able to show the conformational changes upon addition of different ligands and substrates, leading to the proposal that phenol binding may occur on the external side of H55. (4)(5)
Mechanism of Action
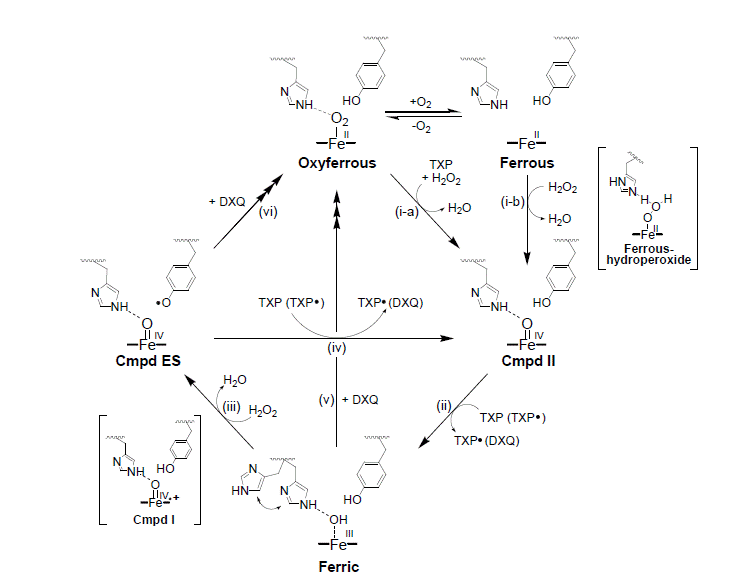
The multi-functional nature of DHP has lead to extensive mechanistic studies. Globins, the oxygen transport function, takes place via an Fe(II) ferrous/oxyferrous heme species. However, peroxidase and peroxygenase activities take place through ferric, Fe(III) species. It has been found that phenolic substrates, such as trihalophenols, act through a radical mechanism, typical of peroxidases and restores DHP to it's native globin function. (3)
Monohalophenols inhibit the active site, preventing and/or hindering substrate turnover. In the case with a great excess of hydrogen peroxide or without substrate, the enzyme goes to an unknown species called compound RH. This species has spectroscopic details of a 'chewed up heme' with bleaching of the soret band, and in most cases, nonrecoverable. However, in the reaction with indole as substrate (and under certain conditions with trichlorophenol), it has been observed that with excess substrate, compound RH will recover/react to turn over to the oxyferrous state. Further studies are being done to determine the structure and chemical composition of compound RH. (1)
The metacyano form of DHP (Fe-CN) shows distinct properties with a strong blue shift in the soret and unique visible band properties. DHP-CO complex also exhibits very distinct Uv-vis features with an increase in the soret and blue shift. Mechanistic studies have been done to determine the functionality of these ligands, but have been unrecoverable because of the strong field nature of these ligands. In current work with using O2 as co-substrate, in order to end the reaction, we have added KCN to the solution which immediately shuts down the reaction by binding CN and leaving the enzyme 'mute' (4)(7).
Why Study Dehaloperoxidase?
There are persistent chemical pollutants including herbicides, pesticides and industrial by-products, among others that pose significant health risks. So, researchers have posed a simple question, can we use DHP for bioremediation purposes? Moreover, DHP has shown promise in possible remediation with Agent Orange (dioxin) that still, decades later, poisons the land and people in Vietnam from a lack of means for proper and safe disposal. Trichlorophenol (TCP) is very toxic analog of dioxin; DHP has effectively and efficiently oxidized and detoxified this compound (1). Heme containing peroxidases have been known for their ability for bioremediation and have been extensively studied (8). However, The analysis of the primary sequence of DHP fails to predict both peroxidase and globin activities. The structural fold of this protein only predicts its globin function; Therefore, DHP challenges our understanding of protein structure-function relationships.
References
(1) D’Antonio, J.; D’Antonio, E. L.; Thompson, M. K.; Bowden, E. F.; Franzen, S.; Smirnova, T.; Ghiladi, R. A.; Spectroscopic and Mechanistic Investigations of Dehaloperoxidase B from Amphitrite ornate. Biochemistry. [Online] 2010, 49, 6600-6616.
(2) D’Antonio, J.; Mechanistic Investigations of Oxyferrous and Ferric States of Dehaloperoxidase B from Amphitrite ornate [dissertation]. [Raleigh (NC)]: North Carolina State University; 2012. p. 2-27, 232.
(3) Franzen, S.; Thompson, M. K.; Ghiladi, R. A.; The dehaloperoxidase paradox. Bio. et. Biophy. Acta [Online] 2012, 1824, 578-588.
(4) Thompson, M. K.; Davis, M. F.; Serrano, V.; Nicoletti, F. P.; Howes, B. D.; Smulevich, G.; Franzen, S.; Internal Binding of Halogenated Phenols in Dehaloperoxidase-Hemoglobin Inhibits Peroxidase Function. Biophysical. [Online] 2010, 99, 1586-1595.
(4) Dunford, B. H.; Heme Peroxidase and Catalase Families. In Peroxidases & Catalases: Biochemistry, Biotechnology, and Physiology. Wiley: Hoboken, New Jersey, 2010; p. 9-12.
(5) Davis, M. F; Structural Studies of Inhibitor and Substrate Binding in Hemoglobin Dehaloperoxidase [dissertation]. [Raleigh (NC)]: North Carolina State University; 2009. p. 3-5, 15-19.
(7) D’Antonio, J.; Ghiladi, R. A.; Reactivity of Deoxy- and Oxyferrous Dehaloperoxidase B from Amphitrite ornate: Identification of Compound II and Its Ferrous-Hydroperoxide Precursor. Biochemistry. [Online] 2011, 50, 5999-6011.
(8) Anastasi, A.; Tigini, V.; Varese, G. C.; The Bioremediation Potential of Different Ecophysiological Groups of Fungi. In Fungi as Bioremediators; Soil Biology. 2013, 32, pp. 29-49.