Aspartate Aminotransferase
From Proteopedia
Aspartate Aminotransferase
by Luke Spooner
Aspartate Aminotransferase (AAT), also known as Glutamic aspartic transaminase, glutamic oxaloacetic transaminase, and transaminase A., is an enzyme that is a member of the class-I pyridoxal-phosphate-dependent aminotransferase family [1]. It is coded by the gene GOT1[2]. It is a homodimer that is 413 amino acids long and serves a critical role in amino acid and carbohydrate metabolism, ureogenesis, and the transfer of reducing equivalents into the mitochondria and chloroplast[3]. Within prokaryote cells it is exclusively found in the cytosol, but in eukaryotic cells there are cytosol, mitochondrial, and chloroplast isozymes[1][4].
In the human body it is produced in the brain, skeletal muscles, liver, pancreas, red blood cells, and kidneys [5][6]. The wide range of tissues in which it is made, separates it from the similar enzyme alanine transaminase (ALT) which is found primarily in the liver[6]. The level of AAT in the body can be used as a marker for tissue disease or damage[6]. As well, AAT and ALT levels can be compared to pinpoint whether tissue damage is primarily found within the liver[6].
Contents |
Structure
|
is a homodimer that contains 16 α-helices and a β-sheet formed from 7 parallel and antiparallel strands[4]. Each subunit contains an equivalent active site[4]. The subunits connect at two sites: between their large domains and between the N-terminal residues and the large domain on the other subunit[7]. This structure of AST varies minutely among organisms ranging from E. coli to humans[4][3]. As well, the structure of the active site is highly conserved with a sequence homology of 25%[4].
Each subunit of the homodimer is further divided into a small and large domain[4]. The is comprised of the amino acids from the N-terminus to Pro 48 residue and from Met 326 to the C-terminus[4]. The remaining amino acids make up the , and the are connected by a long α-helix consisting of 32 amino acids[4].
The large domain is where the active site of AAT is found and to accommodate this, the core contains many α/β supersecondary structures[4]. This is contrasted with the core of the small subunit which is formed from two α-helices and two β-strands[4]. In multicellular organisms there is a kink at the 325th residue which acts as a hinge for the small domain, which allows for the resulting conformational changes that take place upon the binding of inhibitors to the enzyme[4].
As was stated above, the active site of AST is situated on the large domain of the subunit[4]. Within the active site is the amino residue Lys 258, also known as the internal aldimine, which binds with the cofactor Pyridoxal 5'-phosphate () forming what is called a Schiff base[4][3]. Upon the addition of an amino acid substrate, a new Schiiff base forms between PLP and the amino acid[4].
Function
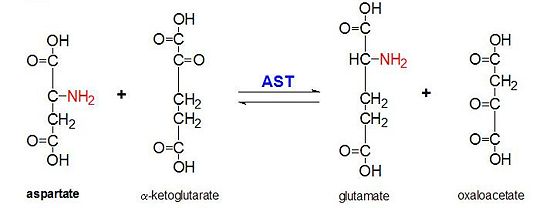
AAT catalyzes the reversible transamination of the α-amino group from L-aspartate to α-ketoglutarate forming oxaloacetate and glutamate[3]. This reactivity is lower in E.coli than in higher eukaryotes, and has broader substrate specificity[4]. However, the reaction takes place in the same way[4]. Upon introduction of an amino acid substrate, a new Schiff base will form between it and the PLP cofactor[4][8]. This causes the amino acid to lose a hydrogen and form a quinoid intermediate, and reprotanation takes place resulting in a ketimine[4][8]. Next, the structure is hydrolyzed forming an α-keto acid and pyridoxamine phosphate[8]. 2-methyl aspartate acts as an inhibitor of AAT when it forms a Schiif base with the PLP cofactor, rather than aspartate[8][4]. This results in the process stopping at the step prior to the alpha protein elimination[8][4].
This reaction is essential to maintaining homeostasis in organisms. The four different molecules that can form as a result of this transanimation (oxaloacetate, α-ketoglutarate, aspartate, L-glutamate) our critical to a number of metabolic processes[9][10][11][3][12]. Oxaloacetate and α-ketoglutarate play a critical role in the Krebs cycle, varying forms of aspartate are important molecules in the urea cycle and participate in gluconeogenesis, and glutamate is an important molecule in metabolic pathways associated with memory[9][10][11][3][12].
Clinical Applications
The levels of AAT in the body are indicative of tissue damage and disease[13]. Normally AAT is found in minimal amounts within the blood, however when the organs mentioned above are damaged, AAT is released into the blood[13]. The amount released is proportional to the level of damage sustained[13]. AAT levels have been shown to rise substantially within 6 hours of the initial tissue degradation and can stay elevated for up to 4 days[13]. AAT levels when compared with the levels of other enzymes can be used by physicians to determine where in the body the damage has taken place[6]. Comparisons with ALT have proven particularly useful in identifying liver damage such as cirrhosis and hepatitis[6]. Under normal condition, AAT levels within men are 6-34 IU/L and for women it is 8-40 IU/L[13].
3D structures of aspartate aminotransferase
Updated on 11-February-2014
1aat – cAAT – chicken
Aspartate aminotransferase binary complex with pyridoxamine phosphate
2aat, 1aia, 1aib, 1aic – EcAAT (mutant) + pyridoxamine phosphate– Escherichia coli
4dbc – EcAAT
9aat - cAAT + pyridoxamine phosphate
1amq, 1amr, 1ams - EcAAT + pyridoxamine phosphate
1a0g - BaAAT + pyridoxamine phosphate – Bacillus
3pd6 - mAAT + pyridoxamine phosphate - mouse
2z9u – MlAAT – Mesorhizobium loti
2z9v – MlAAT + pyridoxamine
Aspartate aminotransferase binary complex with pyridoxal phosphate
1asm, 1asn, 1ars, 1art - EcAAT + pyridoxal phosphate
3aat, 1aam, 1aaw, 1asf, 1asg, 1ahe, 1ahg, 5eaa, 1g4v, 1g4x, 1g7w, 1g7x, 1ix6, 1ix8, 2d65, 2d66, 2d7y, 3zzj, 3zzk, 4a00 - EcAAT (mutant) + pyridoxal phosphate
7aat, 8aat, 1tar, 1tat, 2cst - cAAT + pyridoxal phosphate
1asa, 1asb, 1aka - cAAT (mutant) + pyridoxal phosphate
2dab - BaAAT (mutant) + pyridoxal phosphate
1gc3, 1b5o, 1b5p, 5bj4 - TtAAT (mutant) + pyridoxal phosphate – Thermus thermophilus
1gd9 - PhAAT + pyridoxal phosphate – Pyrococcus horikoshii
1j32 - AAT + pyridoxal phosphate – Phormidium lapideum
1o4s - TmAAT + pyridoxal phosphate – Thermotoga maritima
5bj3 - AAT (mutant) + pyridoxal phosphate – Thermus aquaticus
3k7y - AAT + pyridoxal phosphate – Plasmodium falciparum
3ppl - AAT + pyridoxal phosphate – Corynebacterium glutamicum
2z9w - MlAAT + pyridoxal
3ii0 - AAT + pyridoxal phosphate - human
Aspartate aminotransferase binary complex with pyridoxal phosphate derivative
1ama, 1tas, 1oxo, 1oxp, 1ivr - cAAT + pyridoxal phosphate derivative
1akb, 1akc - cAAT (mutant) + pyridoxal phosphate derivative
1spa, 1asl, 1asd, 1ase, 1arg, 1c9c, 1cq6, 1cq7, 1cq8, 3qn6, 3qpg, 4f5m - EcAAT + pyridoxal phosphate derivative
1asc, 1arh, 1bqa, 1bqd, 1toe, 4f5f, 4f5g, 4f5h, 4f5i, 4f5j, 4f5k, 4f5l - EcAAT (mutant) + pyridoxal phosphate derivative
1x28, 1x29, 1x2a - EcAAT + phosphopyridoxyl glutamate derivative
5daa - BaAAT (mutant) + pyridoxamine phosphate
1maq, 1map – cAAT + ketamine
1ajr, 1ajs - pAAT + pyridoxal phosphate derivative – pig
1bjw - TtAAT + pyridoxal phosphate derivative
2gb3 - TmAAT + pyridoxal phosphate derivative
3meb - AAT + pyridoxal phosphate derivative – Giardia lamblia
3hlm - mAAT + pyridoxal phosphate derivative
3f6t - AAT + pyridoxal phosphate derivative – Lactobacillus acidophilus
4eu1 - AAT + pyridoxal phosphate derivative – Trypanosoma brucei
3nra - AAT + pyridoxal phosphate derivative – Rhodobacter sphaeroides
3lqs - BaAAT + thiophenecarboxylic acid
2z9x - MlAAT + pyridoxyl-alanine
Aspartate aminotransferase ternary complex
1ahf - EcAAT (mutant) + indolylpropionic acid + pyridoxal phosphate
1ahx, 1tog - EcAAT (mutant) + phenylpropionic acid + pyridoxal phosphate
1toi, 1toj - EcAAT (mutant) + phenylpropionic acid + pyridoxal phosphate derivative
1ahy, 1ari, 1qir, 1qis, 1qit, 1b4x, 1ix7, 2d61, 2d7z - EcAAT (mutant) + maleic acid + pyridoxal phosphate
2q7w - EcAAT (mutant) + thiophenecarboxylic acid + pyridoxal phosphate
2qbt, 2qa3, 2qb2, 2qb3 - EcAAT + thiophenecarboxylic acid + pyridoxal phosphate
1tok - EcAAT (mutant) + maleic acid + pyridoxal phosphate derivative
1czc - EcAAT (mutant) + glutaric acid + pyridoxal phosphate
1cze - EcAAT (mutant) + succinic acid + pyridoxal phosphate
1yoo, 2d5y, 2d63, 2d64 - EcAAT (mutant) + isovaleric acid + pyridoxal phosphate
3pa9, 3paa - EcAAT + furancarboxylic acid + pyridoxamine phosphate
1gc4, 1gck - TtAAT (mutant) + aspartic acid + pyridoxal phosphate
1gc3 - TtAAT (mutant) + tryptophan + pyridoxal phosphate
1bkg - TtAAT + maleic acid + pyridoxamine phosphate
1gde - PhAAT + glutamic acid + pyridoxal phosphate
1yaa - yAAT + maleic acid + pyridoxal phosphate – yeast
3pdb - mAAT + oxaloacetic acid + pyridoxamine phosphate
References
- ↑ 1.0 1.1 Han Q, Robinson H, Cai T, Tagle DA, Li J. Biochemical and structural characterization of mouse mitochondrial aspartate aminotransferase, a newly identified kynurenine aminotransferase-IV. Biosci Rep. 2010 Oct 26. PMID:20977429 doi:10.1042/BSR20100117
- ↑ DeLorenzo RJ, Ruddle FH. Glutamate oxalate transaminase (GOT) genetics in Mus musculus: linkage, polymorphism, and phenotypes of the Got-2 and Got-1 loci. Biochem Genet. 1970 Apr;4(2):259-73. PMID:4193185
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Jeffery CJ, Gloss LM, Petsko GA, Ringe D. The role of residues outside the active site: structural basis for function of C191 mutants of Escherichia coli aspartate aminotransferase. Protein Eng. 2000 Feb;13(2):105-12. PMID:10708649
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 4.18 4.19 Kamitori S, Okamoto A, Hirotsu K, Higuchi T, Kuramitsu S, Kagamiyama H, Matsuura Y, Katsube Y. Three-dimensional structures of aspartate aminotransferase from Escherichia coli and its mutant enzyme at 2.5 A resolution. J Biochem. 1990 Aug;108(2):175-84. PMID:2121725
- ↑ Palaiologos G, Hertz L, Schousboe A. Role of aspartate aminotransferase and mitochondrial dicarboxylate transport for release of endogenously and exogenously supplied neurotransmitter in glutamatergic neurons. Neurochem Res. 1989 Apr;14(4):359-66. PMID:2569674
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 Tran A, Longo F, Ouzan D, Bianchi D, Pradier C, Saint-Paul MC, Sattonnet C, Laffont C, Dantin S, Piche T, Benzaken S, Rampal P. Effects of 1-year interferon-alpha 2a treatment in patients with chronic hepatitis C and persistently normal transaminase activity. Scand J Gastroenterol. 2000 Apr;35(4):433-7. PMID:10831269
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedAAT_Structure - ↑ 8.0 8.1 8.2 8.3 8.4 Martinez-Carrion M, Tiemeier DC, Peterson DL. Conformational properties of the isoenzymes of aspartate transaminase and the enzyme-substrate complexes. Biochemistry. 1970 Jun 23;9(13):2574-82. PMID:5450225
- ↑ 9.0 9.1 Tretter L, Adam-Vizi V. Inhibition of Krebs cycle enzymes by hydrogen peroxide: A key role of [alpha]-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. J Neurosci. 2000 Dec 15;20(24):8972-9. PMID:11124972
- ↑ 10.0 10.1 Tretter L, Adam-Vizi V. Inhibition of Krebs cycle enzymes by hydrogen peroxide: A key role of [alpha]-ketoglutarate dehydrogenase in limiting NADH production under oxidative stress. J Neurosci. 2000 Dec 15;20(24):8972-9. PMID:11124972
- ↑ 11.0 11.1 Jungas RL, Halperin ML, Brosnan JT. Quantitative analysis of amino acid oxidation and related gluconeogenesis in humans. Physiol Rev. 1992 Apr;72(2):419-48. PMID:1557428
- ↑ 12.0 12.1 Gibbs ME, Hertz L. Importance of glutamate-generating metabolic pathways for memory consolidation in chicks. J Neurosci Res. 2005 Jul 15;81(2):293-300. PMID:15929064 doi:10.1002/jnr.20548
- ↑ 13.0 13.1 13.2 13.3 13.4 Gonzalez-Flecha B, Cutrin JC, Boveris A. Time course and mechanism of oxidative stress and tissue damage in rat liver subjected to in vivo ischemia-reperfusion. J Clin Invest. 1993 Feb;91(2):456-64. PMID:8432855 doi:http://dx.doi.org/10.1172/JCI116223