User:A. Rahim Zalal/Sandbox 1
From Proteopedia
Histone (H2A)
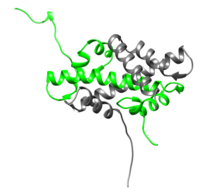
The histone family's H2A protein family act as a subunits of eukaryotic nucleosomes. Specifically, H2A molecules form dimers with H2B molecules that interact with tetramers of histones H3 and H4 to form the octameric histone core of the nucleosome.[1] All of the core histone proteins, including H2A, also interact with DNA in ways that allow for DNA compaction.
Contents |
Background
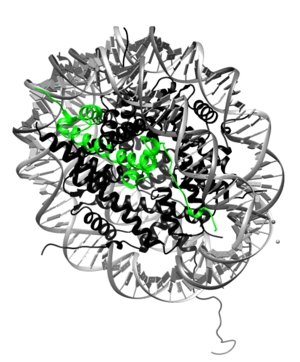
The histone proteins are a family of proteins that allow for the compaction of DNA into functional units known as nucleosomes, which, through certain organized arrangements, form the chromatin fibers in eukaryotic organisms. The histone family includes the core histone families, H2A (shown highlighted on top right), H2B, H3, and H4, as well as the linker histone family, H1. The core histone proteins form the core of the nucleosome, while the linker histones sit on the outside of the nucleosome subunit and assist in interactions with other nucleosomes within the chromatin. To form the core, H3 and H4 form a tetramer that is then joined by two H2A:H2B dimers. This results in two of each core protein forming a histone octamer. The core allows for about 1.7 loops of DNA to encircle it. The role of histone proteins in the genetics of an organism also provides another avenue to study the effects of histone protein modifications on the epigenetics of the organism in question.[2]
Structure
| |||||||||||
Function
Sequence
Protein Primary Sequence
The sequence for histone H2A type 1 (human) is presented here[4]:
10 20 30 40 50 60
MSGRGKQGGK ARAKAKTRSS RAGLQFPVGR VHRLLRKGNY AERVGAGAPV YLAAVLEYLT
70 80 90 100 110 120
AEILELAGNA ARDNKKTRII PRHLQLAIRN DEELNKLLGK VTIAQGGVLP NIQAVLLPKK
130
TESHHKAKGK
Coding DNA Sequence
The coding DNA sequence for the H2A type 1 (human)[5]:
ENA|AAA63191|AAA63191.1 Homo sapiens (human) histone H2A.1 : Location:1..393 ATGTCTGGACGTGGAAAGCAAGGCGGCAAAGCTCGGGCAAAAGCTAAAACGCGTTCTTCC AGGGCCGGTCTTCAGTTTCCAGTTGGCCGTGTGCACCGCCTCCTCCGCAAAGGCAACTAC TCCGAACGAGTCGGGGCCGGCGCTCCAGTGTACCTGGCAGCGGTGCTGGAATATCTGACG GCCGAGATCTTAGAGCTAGCTGGCAACGCGGCTCGCGACAATAAGAAGACCCGCATCATC CCGCGCCACCTGCAGCTAGCCATCCGCAACGACGAGGAGCTAAATAAGCTTCTAGGTCGC GTGACCATCGCGCAGGGCGGTGTCCTGCCCAACATCCAGGCCGTATTGCTGCCTAAGAAG ACGGAGAGCCACCATAAGGCCAAGGGCAAGTGA
Mechanism
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
References
- ↑ Rhodes D. Chromatin structure. The nucleosome core all wrapped up. Nature. 1997 Sep 18;389(6648):231, 233. PMID:9305832 doi:http://dx.doi.org/10.1038/38386
- ↑ Kass SU, Wolffe AP. DNA methylation, nucleosomes and the inheritance of chromatin structure and function. Novartis Found Symp. 1998;214:22-35; discussion 36-50. PMID:9601010
- ↑ Rhodes D. Chromatin structure. The nucleosome core all wrapped up. Nature. 1997 Sep 18;389(6648):231, 233. PMID:9305832 doi:http://dx.doi.org/10.1038/38386
- ↑ Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics. 2002 Nov;80(5):487-98. PMID:12408966
- ↑ Albig W, Kardalinou E, Drabent B, Zimmer A, Doenecke D. Isolation and characterization of two human H1 histone genes within clusters of core histone genes. Genomics. 1991 Aug;10(4):940-8. PMID:1916825