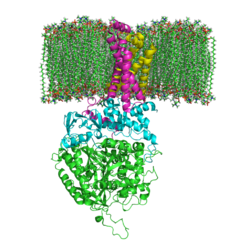
Succinate Dehydrogenase
From Proteopedia
| |||||||||||
Contents |
Mechanisms
Succinate oxidation
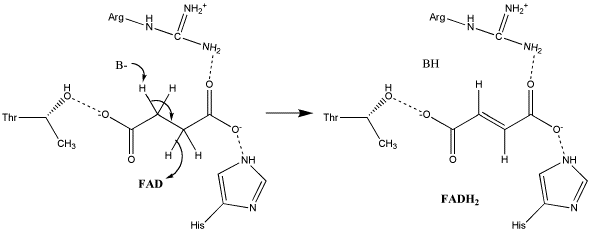
The exact mechanism for the oxidation of succinate to fumarate has not yet been elucidated. The initial deprotonation may be performed by FAD, Glu255, Arg286, or His242 of SdhA, and the following elimination may be a concerted E2 or E1cb elimination. In the concerted mechanism, the α-carbon is deprotonated by a base as FAD removes a hydride from the β-carbon; this is shown in image 1 [9].
Image 1: Oxidation of succinate to fumarate through E2 elimination (from Adamandalex in Wikimedia Commons http://en.wikipedia.org/wiki/File:S.D.Oxidation_of_Succinate_E2.gif).
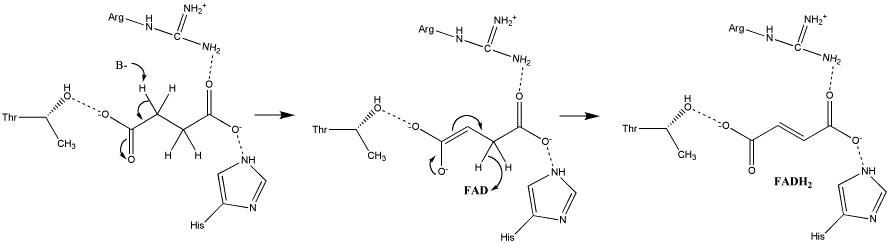
In the proposed E1cb mechanism, the deprotonation leads to the formation of an enolate intermediate; FAD then removes the hydride, as shown in Image 2 [9].
Image 2: Oxidation of succinate to fumarate via E1cb elimination (from Adamandalex in Wikimedia Commons http://en.wikipedia.org/wiki/File:S.D.Oxidation_of_Succinate_E1cb.gif).
Ubiquinone reduction
Ubiquinone is initially oriented in the active site such that the O1 carbonyl group interacts with Tyr83 of SdhD via hydrogen bonding. The electrons removed during the oxidation reaction are conveyed through the iron-sulfur clusters to 3Fe-4S; their presence on that cluster stimulates the substrate to reorient so that a second hydrogen bond between the of SdhC may form. The electrons are transferred to the substrate individually, with the addition of the first producing a radical semiquinone and the second completing the reduction to ubiquinol. This mechanism is illustrated in image 3 [9].
Image 3: Reduction of ubiquinone to ubiquinol (from Adamandalex in Wikimedia Commons http://en.wikipedia.org/wiki/File:QuinoneMechanism.gif).
Regulation
Since succinate dehydrogenase possesses multiple active sites that catalyze two different reactions, two classes of inhibitors function on the enzyme. The first class, which includes succinate analogs--both naturally-occuring TCA cycle intermediates like malate and oxaloacetate and the synthetic analog, malonate--contains some of the strongest succinate dehydrogenase inhibitors. The second class of inhibitors, which includes the ubiquinone analogs thenoyltrifluoroacetone and carboxin, binds to the ubiquinone active site and prevents reduction of the substrate[10].
3D structures of succinate dehydrogenase
Updated on 24-September-2014
SDH
2wdv - EcSDH flavoprotein + Fe-S protein + cytochrome B-556 + membrane anchor protein subunits – Escherichia coli
2wp9 - EcSDH flavoprotein + Fe-S protein (mutant) + cytochrome B-556 + membrane anchor protein subunits
2ws3, 2wu2, 2wu5 - EcSDH flavoprotein + Fe-S protein + cytochrome B-556 + membrane anchor protein subunits (mutant)
3aef, 1zoy - pSDH flavoprotein + Fe-S protein + cytochrome B subunits - pig
3vra, 3vr8 - prSDH flavoprotein + Fe-S protein + cytochrome B subunits – pig roundworm
2h88 - cSDH flavoprotein + IP + cytochrome B subunits - chicken
2lm4 – SDH assembly factor subunit – yeast – NMR
SDH binary complexes
1nek – EcSDH flavoprotein + Fe-S protein + cytochrome B-556 + membrane anchor protein subunits + ubiquinone
1nen - EcSDH flavoprotein + Fe-S protein + cytochrome B-556 + membrane anchor protein subunits + dinitrophenol-17
2acz - EcSDH flavoprotein + Fe-S protein + cytochrome B-556 + membrane anchor protein subunits + atpenin
2wdq - EcSDH flavoprotein + Fe-S protein + cytochrome B-556 + membrane anchor protein subunits + carboxin
2wdr - EcSDH flavoprotein + Fe-S protein + cytochrome B-556 + membrane anchor protein subunits + pentachlorophenol
3abv, 3ae1, 3ae2, 3ae3, 3ae4, 3ae5, 3ae6, 3ae7, 3ae8, 3ae9, 3aea, 3aeb, 3aec, 3aed, 3aeg - pSDH flavoprotein + Fe-S protein + cytochrome B subunits + benzamide derivative
3aee - pSDH flavoprotein + Fe-S protein + cytochrome B subunits + atpenin
2fbw, 2wqy - cSDH flavoprotein + IP + cytochrome B subunits + carboxin
2h89 - cSDH flavoprotein + IP + cytochrome B subunits + malonate
3vr9 - prSDH flavoprotein + Fe-S protein + cytochrome B subunits + flutolanil
SDH ternary complexes
3sfd - pSDH flavoprotein + Fe-S protein + cytochrome B subunits + oxalacetate + pentachlorophenol
3sfe - pSDH flavoprotein + Fe-S protein + cytochrome B subunits + oxalacetate + thiabendazole
1zp0 - pSDH flavoprotein + Fe-S protein + cytochrome B subunits + thenoyltrifluoroactone + nitropropionate
1yq3 - cSDH flavoprotein + IP + cytochrome B subunits + ubiquinone + oxalacetate
1yq4 - cSDH flavoprotein + IP + cytochrome B subunits + ubiquinone + nitropropionate
3vrb - prSDH flavoprotein + Fe-S protein + cytochrome B subunits + flutolanil + fumarate
References
- ↑ Oyedotun KS, Lemire BD. The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase. Homology modeling, cofactor docking, and molecular dynamics simulation studies. J Biol Chem. 2004 Mar 5;279(10):9424-31. Epub 2003 Dec 12. PMID:14672929 doi:10.1074/jbc.M311876200
- ↑ Tomitsuka E, Hirawake H, Goto Y, Taniwaki M, Harada S, Kita K. Direct evidence for two distinct forms of the flavoprotein subunit of human mitochondrial complex II (succinate-ubiquinone reductase). J Biochem. 2003 Aug;134(2):191-5. PMID:12966066
- ↑ 3.0 3.1 Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003 Jan 31;299(5607):700-4. PMID:12560550 doi:10.1126/science.1079605
- ↑ 4.0 4.1 Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, Byrne B, Cecchini G, Iwata S. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J Biol Chem. 2006 Mar 17;281(11):7309-16. Epub 2005 Dec 27. PMID:16407191 doi:http://dx.doi.org/10.1074/jbc.M508173200
- ↑ Kenney WC. The reaction of N-ethylmaleimide at the active site of succinate dehydrogenase. J Biol Chem. 1975 Apr 25;250(8):3089-94. PMID:235539
- ↑ Voet, Donald, Charlotte W. Pratt, and Judith G. Voet. Fundamentals of Biochemistry: Life at the Molecular Level. 2nd Ed. Hoboken, NJ: Wiley, 2008.
- ↑ Vinogradov AD, Kotlyar AB, Burov VI, Belikova YO. Regulation of succinate dehydrogenase and tautomerization of oxaloacetate. Adv Enzyme Regul. 1989;28:271-80. PMID:2624174
- ↑ Boyd AW, Lackmann M. Signals from Eph and ephrin proteins: a developmental tool kit. Sci STKE. 2001 Dec 11;2001(112):re20. PMID:11741094 doi:10.1126/stke.2001.112.re20
- ↑ 9.0 9.1 9.2 Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b. J Biol Chem. 2006 Oct 27;281(43):32310-7. Epub 2006 Sep 1. PMID:16950775 doi:10.1074/jbc.M607476200
- ↑ Muller FL, Liu Y, Abdul-Ghani MA, Lustgarten MS, Bhattacharya A, Jang YC, Van Remmen H. High rates of superoxide production in skeletal-muscle mitochondria respiring on both complex I- and complex II-linked substrates. Biochem J. 2008 Jan 15;409(2):491-9. PMID:17916065 doi:10.1042/BJ20071162
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Michael Vick, David Canner, Alexander Berchansky