From Proteopedia
proteopedia linkproteopedia link |
Introduction
Discovered and isolated by Anselme Payen in 1833, amylase was the first enzyme to be discovered[1]. Amylases are hydrolases, acting on α-1,4-glycosidic bonds[2]. They can be further subdivided into α,β and γ amylases[1].α-Amylase (AAM) is an enzyme that acts as a catalyst for the hydrolysis of alpha-linked polysaccharides into α-anomeric products[3]. The enzyme can be derived from a variety of sources, each with different characteristics. α-Amylase found within the human body serves as the enzyme active in pancreatic juice and salvia[2]. α-Amylase is not only essential in human physiology but has a number of important biotechnological functions in various processing industries. Beta/alpha amylase (BAAM) is a precursor protein which is cleaved to form the beta-amylase and alpha-amylase after secretion.
Structure[3]
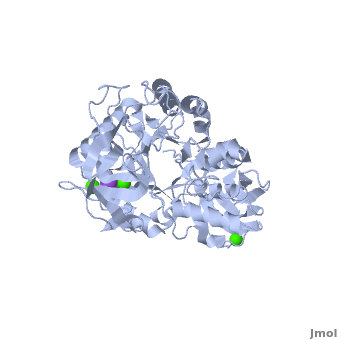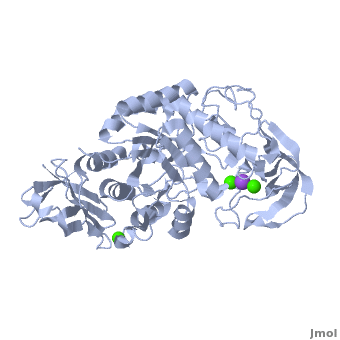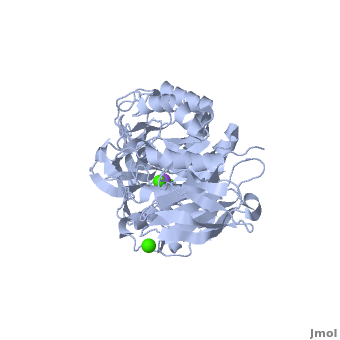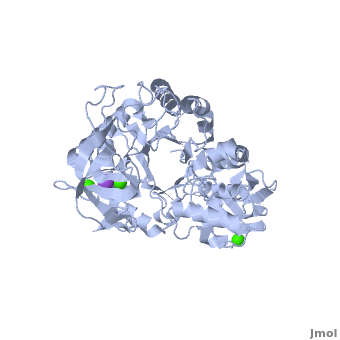
Shown as 1hvx is the structure of the thermostable α-amylase of Bacillus stearothermophilus (BSTA)[3]. BSTA is comprised of a single polypeptide chain. This chain is folded into three domains: A, B and C. These domains are generally found on all α-amylase enzymes. The constitutes the core structure, with a (β/α)8-barrel.The consists of a sheet of four anti-parallel β-strands with a pair of anti-parallel β-strands. Long loops are observed between the β-strands. Located within the B domain is the for Ca2+-Na+-Ca2+. consisting of eight β-strands is assembled into a globular unit forming a Greek key motif. It also holds the Ca2+ binding site in association with domain A. Positioned on the C-terminal side of the β-strands of the (β/α)8-barrel in domain A is the active site. The catalytic residues involved for the BSTA active site are
. The residues are identical to other α-amylases, yet there are positional differences which reflect the flexible nature of catalytic resides.
found in the interior of domain B and at the interface of domain A and C, constitute the metal ion binding sites. All α-amylases contain one strongly conserved Ca2+ ion for structural integrity and enzymatic activity.[4] CaI is consistent in α-amylases, however there are structural differences between the linear trio of CaI, CaII and Na in other enzymes. CaIII acts as a bridge between two loops, one from α6 of domain A, and one between β1 and β2 of domain C.
Chloride Dependent Enzymes
A family of chloride-dependent enzymes, including salivary and pancreatic α-amylase, require the binding of a chloride ion to be allosterically activated[4]. The function of the chloride ion still remains uncertain. No relationship has been observed between the anion binding affinity and its activity, indicating the complexity between the binding parameters and mechanism it activates[4]. Studies have shown that nitrite and nitrate ions with pancreatic α-amylase fit within the chloride binding site, thus making all the necessary hydrogen bonds and enhancing the relative activity by 5-fold[5].
Function
Mechanism
In the human body, α-amylase is part of digestion with the breakdown of carbohydrates in the diet. The mechanism involved includes catalyzing substrate hydrolysis by a double replacement mechanism, forming a covalent glycosyl-enzyme intermediate and hydrolyzed through oxocarbenium ion-like transition states[6]. One of the carboxylic acids in the active site acts as the catalytic nucleophile during the formation of the intermediate. A second carboxylic acid operates as the acid/base catalyst, supporting the stabilization of the transition states during the hydrolysis[6].
Human Salivary and Pancreatic α-Amylase
Salivary α-Amylase hydrolyzes the (α1-4) glycosidic linkages of starch, separating it into short polysaccharide fragments[7]. Once the enzyme reaches the stomach, it becomes inactivated due to the acidic pH. Further breakdown of starch occurs by secretion of a second form of the enzyme by the pancreas. Pancreatic juice enters the duodenum and pancreatic α-amylase further cleaves starch to yield maltose, maltotriose and oligosaccharides[7]. The oligosaccharides are referred to as dextrins, which are fragments of amylopectin consisting of (α1-6)branch points[7]. Microvilli of the intestinal epithelia break maltose and dextrins into glucose, which gets absorbed into the circulatory system[7]. Glycogen has a relatively similar structure as starch, and thus proceeds in the same digestive pathway.
Regulation
α-Amylase is regulated through a number of inhibitors. These inhibitors are classified according to six categories, based on their tertiary structures[8]. Inhibitors of α-amylase block the active site of the enzyme. In animals, inhibitors control the conversion of starch to simple sugars during glucose peaks after a meal so that breakdown of glucose occurs at a rate the body can handle[8]. This is particularly important for diabetics, who require low quantities of α-amylase to maintain control over glucose levels. After taking insulin however, pancreatic α-amylase escalates. Plants use these inhibitors as a defense mechanism to inhibit the use of α-amylase in insects, thus protecting themselves from herbivory[9].
Industrial Uses
α-Amylase is used extensively in various industrial processes. In textile weaving, starch is added for warping. After weaving, the starch is removed by Bacillus subtilis α-amylase[1]. Dextrin, which is a viscosity improver, filler, or ingredient of food, is manufactured by the liquefaction of starch by bacteria α-amylase[1]. Bacterial α-amylases of B.subtilis, or B.licheniformis are used for the initial starch liquefaction in producing high conversion glucose syrup[1]. Pancreatitis can be tested by determining the level of amylases in the blood, a result of damaged amylase-producing cells, or excretion due to renal failure[10]. α-Amylase is used for the production of malt, as the enzyme is produced during the germination of cereal grains[1].
β/α amylase (BAAM) is a precursor protein which is cleaved to form the β-amylase and α-amylase after secretion.
|
3D structures of ricin ( Updated on 28-October-2014 )
(( 1ua7 – BsAAM + acarbose
Pullulanase α-amylase binary complexes
2fh6, 2fh8, 2fhb, 2fhc, 2fhf - KaAAM + saccharide
3fax - AAM + saccharide – Streptococcus agalactiae
2e8z, 2e9b - BsAAM + saccharide
2d2o - TvAAM + saccharide
1g1y, 1jib, 1jl8, 1vfm, 1vfo, 1vfu, 1vb9 - TvAAM (mutant) + saccharide
3a6o – TvAAM + acarbose
Neopullulanase α-amylase
4aef – AAM – Pyrococcus furiosus
1j0h – BsAAM
2z1k – AAM – Thermus thermophilus
1j0i – BsAAM + α-D-glucose
1j0k – BsAAM (mutant) + α-D-glucose
1j0j – BsAAM (mutant) + maltotetraose
β-amylase
2xfr – bBAM
1wdp – sBAM – soybean
2dqx, 1uko, 1ukp – sBAM (mutant)
1vem, 5bca, 1cqy, 1b90 – BcBAM – Bacillus cereus
1ven - BcBAM (mutant)
1fa2 - AAM + saccharide – Sweet potato
β-amylase binary complexes
2xff – bBAM + acarbose
2xfy, 2xg9, 2xgb, 2xgi – bBAM + inhibitor
1wdq, 1wdr, 1wds, 1v3h, 1v3i, 1q6d, 1q6e, 1q6f, 1q6g - sBAM (mutant) + saccharide
1q6c, 1bfn, 1bya, 1byb, 1byc, 1byd, 1btc - sBAM + saccharide
1j0y, 1j0z, 1j10, 1j11, 1j12, 1j18, 1b9z - BcBAM + saccharide
1veo, 1vep, 1itc - BcBAM (mutant) + saccharide
1b1y - AAM (mutant) + saccharide – Hordeum vulgare
γ-amylase
1lf6 – TtGAM – Thermoanaerobacterium thermosaccharolyticum
1lf9 - TtGAM + acarbose
β/α-amylase
2laa, 2lab – PpBAAM – Paenibacillus polymyxa - NMR
3voc – PpBAAM
Maltohexaose-producing amylase
1wp6 - BacMAM
1wpc – BacMAM + saccharide
Maltogenic amylase
1gvi, 1sma – MAM – Thermus sp.
Taka amylase
2taa, 3vx0, 3vx1 – AoTAM
7taa – AoTAM + acarbose
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Yamamoto T.1988. Handbook of Amylases and Related Enzymes: Their Sources, Isolation Methods, Properties and Applications. Osaka Japan: Pergamon Press
- ↑ 2.0 2.1 Aghajari N, Feller G, Gerday C, Haser R. Crystal structures of the psychrophilic alpha-amylase from Alteromonas haloplanctis in its native form and complexed with an inhibitor. Protein Sci. 1998 Mar;7(3):564-72. PMID:9541387
- ↑ 3.0 3.1 3.2 Suvd D, Fujimoto Z, Takase K, Matsumura M, Mizuno H. Crystal structure of Bacillus stearothermophilus alpha-amylase: possible factors determining the thermostability. J Biochem. 2001 Mar;129(3):461-8. PMID:11226887
- ↑ 4.0 4.1 4.2 Aghajari N, Feller G, Gerday C, Haser R. Structural basis of alpha-amylase activation by chloride. Protein Sci. 2002 Jun;11(6):1435-41. PMID:12021442
- ↑ Maurus R, Begum A, Williams LK, Fredriksen JR, Zhang R, Withers SG, Brayer GD. Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity(,). Biochemistry. 2008 Mar 18;47(11):3332-44. Epub 2008 Feb 20. PMID:18284212 doi:10.1021/bi701652t
- ↑ 6.0 6.1 Maurus R, Begum A, Williams LK, Fredriksen JR, Zhang R, Withers SG, Brayer GD. Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity(,). Biochemistry. 2008 Mar 18;47(11):3332-44. Epub 2008 Feb 20. PMID:18284212 doi:10.1021/bi701652t
- ↑ 7.0 7.1 7.2 7.3 Kuriki T, Imanaka T. The concept of the alpha-amylase family: structural similarity and common catalytic mechanism. J Biosci Bioeng. 1999;87(5):557-65. PMID:16232518
- ↑ 8.0 8.1 PPMID: 17713601
- ↑ Franco OL, Rigden DJ, Melo FR, Grossi-De-Sa MF. Plant alpha-amylase inhibitors and their interaction with insect alpha-amylases. Eur J Biochem. 2002 Jan;269(2):397-412. PMID:11856298
- ↑ Yang RW, Shao ZX, Chen YY, Yin Z, Wang WJ. Lipase and pancreatic amylase activities in diagnosis of acute pancreatitis in patients with hyperamylasemia. Hepatobiliary Pancreat Dis Int. 2005 Nov;4(4):600-3. PMID:16286272