1ygp
From Proteopedia
| |||||||
| , resolution 2.8Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | and | ||||||
| Gene: | YEAST GLYCOGEN PHOSPHORYLASE (Saccharomyces cerevisiae) | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
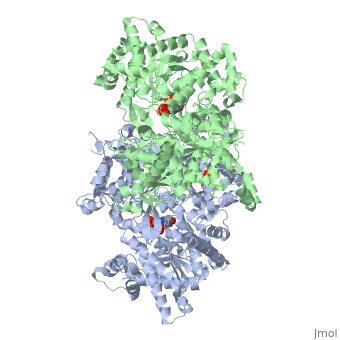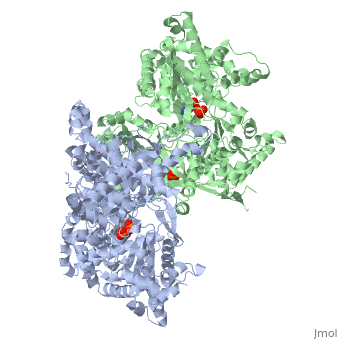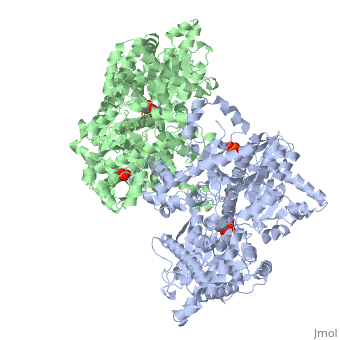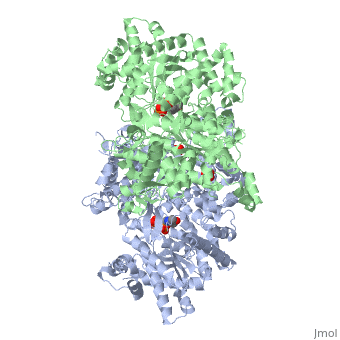
PHOSPHORYLATED FORM OF YEAST GLYCOGEN PHOSPHORYLASE WITH PHOSPHATE BOUND IN THE ACTIVE SITE.
Overview
A phosphorylation-initiated mechanism of local protein refolding activates yeast glycogen phosphorylase (GP). Refolding of the phosphorylated amino-terminus was shown to create a hydrophobic cluster that wedges into the subunit interface of the enzyme to trigger activation. The phosphorylated threonine is buried in the allosteric site. The mechanism implicates glucose 6-phosphate, the allosteric inhibitor, in facilitating dephosphorylation by dislodging the buried covalent phosphate through binding competition. Thus, protein phosphorylation-dephosphorylation may also be controlled through regulation of the accessibility of the phosphorylation site to kinases and phosphatases. In mammalian glycogen phosphorylase, phosphorylation occurs at a distinct locus. The corresponding allosteric site binds a ligand activator, adenosine monophosphate, which triggers activation by a mechanism analogous to that of phosphorylation in the yeast enzyme.
About this Structure
1YGP is a Single protein structure of sequence from Saccharomyces cerevisiae. The following page contains interesting information on the relation of 1YGP with [Glycogen Phosphorylase]. Full crystallographic information is available from OCA.
Reference
A protein phosphorylation switch at the conserved allosteric site in GP., Lin K, Rath VL, Dai SC, Fletterick RJ, Hwang PK, Science. 1996 Sep 13;273(5281):1539-42. PMID:8703213
Page seeded by OCA on Thu Mar 20 15:22:42 2008