Introduction
FAS-II System
Mechanism of Action
General Structural Information
Crystal structures of InhA reveal a (each subunit featured with a different color) in aqueous solution with separate ligand binding sites in each subunit. Each subunit is composed of 289 residues and features a typical
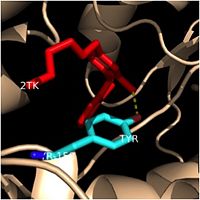
[1] containing a single NADH binding site. The of InhA is made up of several alpha helices (pink), beta sheets (gold), and beta turns (white). This enzyme also features a fatty acyl binding crevice that accommodates the long-chain fatty acyl substrate (2TK) needed to synthesize mycolic acid precursors. The of the InhA form one side of the fatty acyl binding crevice, referred to as the (residues 196-219).
Fatty Acyl Binding Crevice (substrate binding loop in purple; substrates pictured inside the crevice)
Talk generally about catalytic triad. More specific details in sections below.
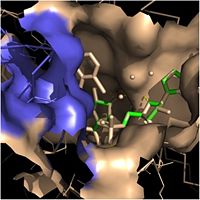
Fatty Acyl Binding Crevice
Within the fatty acyl binding crevice, the NADH substrate sits on the top shelf of the Rossmann fold, and the fatty acyl substrate sits on top of the NADH substrate. Both of these substrates are held in place through interactions with the hydrophobic residues in the substrate binding loop.
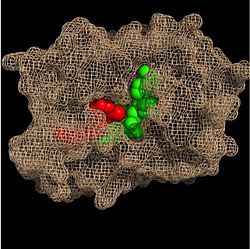
Substrate Binding Pocket (NADH in green; fatty acyl substrate in red)
One side of this crevice is open and exposed to solvent, which allows the substrates to access the binding pocket of this enzyme. The size of the substrate binding loop is a primary determinant of the ability of InhA to distinguish between shorter and longer substrates and provide for the enoyl-ACP reductase reaction.
Catalytic Triad
Hydrogen Bonding Interactions
.
Clinical Applications
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.