User:Braden Sciarra/Sandbox 1
From Proteopedia
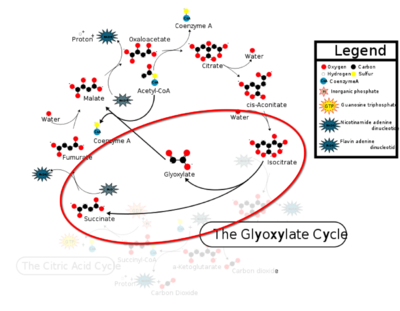
Isocitrate Lyase from Mycobacterium Tuberculosis
| |||||||||||
References
- ↑ Sharma V, Sharma S, Hoener zu Bentrup K, McKinney JD, Russell DG, Jacobs WR Jr, Sacchettini JC. Structure of isocitrate lyase, a persistence factor of Mycobacterium tuberculosis. Nat Struct Biol. 2000 Aug;7(8):663-8. PMID:10932251 doi:10.1038/77964
- ↑ . The interlocking mechanism created by these helices provides additional strength to hold the two monomeric subunits together, allowing ICL to essentially be composed of two dimerized subunits. This interaction will bury approximately 18% of the surface of each subunit, and will help to shield the interior binding site from hydration.
Active Site
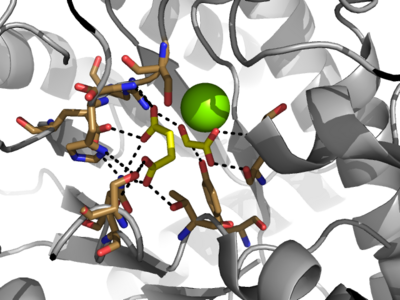
The of isocitrate lyase lies near the C-terminal ends of the Beta-strands of the active site. The glyoxylate substrate is held into place by a network of hydrogen bonds with Ser 91, Gly 92, Trp 93, and Arg 228<ref> <li id="cite_note-2">[[#cite_ref-2|↑]] . The ordered hydrogen bonding within the active site orients the succinate molecule such that the alpha carbon is only 3.2 angstroms away from the deprotonated thiol group of Cys 191<ref name="ICL"</li> <li id="cite_note-3">[[#cite_ref-3|↑]] . This closed conformation will cause the binding site to become inaccessible to the solvent. The loop closure is triggered by the movement of the Mg2+ ion that occurs upon binding of the succinate. This movement of the Mg2+ ion results in electrostatic interactions at LYS189, causing the loop to close.
==Mechanism== [[Image:ICL Mechanism.png|400 px|right|thumb|Figure 4: Chemical Mechanism of Isocitrate Lyase]] Isocitrate lyase catalyzes a reversible aldol condensation, converting isocitrate to glyoxylate and succinate via the breaking of a C-C bond. Within the active site of ICL the HIS193 residue deprotonates the CYS191 residue of the active site in order to increase its basicity<ref name="ICL"</li> <li id="cite_note-4">[[#cite_ref-4|↑]] . The protonation of this species will yield the final product. It is important to note that this reaction is entirely reversible; the breakdown of isocitrate into glyoxylate and succinate occurs using a similar mechanism.
==Elucidation of ICL Structure Using Inhibitors== [[Image:Inhibitors57.png|400 px|right|thumb|Figure 5: Substrate inhibitors of Isocitrate Lyase]] The two inhibitors used to elucidate the structure of ICL were 3-nitropropionate and 3-bromopyruvate. The 3-nitropropionate was used to mimic the succinate, while the 3-bromopyruvate is used to mimic the glyoxylate. These two inhibitors have also been shown to be good inhibitors of isocitrate lyase in ''M. avium'' indicating that their inhibitory capacity is conserved across multiple species. A mutant isocitrate lyase C191S, in conjunction with the aforementioned substrate mimics, was used to elucidate the first high resolution crystal structure of ICL. Dehalogenated 3-nitropropionate works to inhibit isocitrate lyase by forming a covalent bond with the SER191 residue in the active site. This 3-nitropropionate occupies the same site that the succinate would occupy. The C191S mutant adopts a conformation almost identical to the CYS191 residue in the wild type indicating that this is an accurate depiction of the conformation.
</li></ol></ref>