Introduction
Mycobacterium Tuberculosis is an ACSVL (Acyl-CoA synthetases very long) peripheral membrane protein[1]. ACS proteins activate lipids and fatty acids before going into metabolic pathways. FadD13 is soluble unlike other ACSVL proteins. FadD13 contains a hydrophobic tunnel for fatty acids to bind to, as well as an arginine rich lid loop that binds to the cell membrane. The binding of ATP causes structural changes promoting the binding of the hydrophobic substrates. Formation of an acyl-adenylate intermediate induces a 140 degree rotation of the small domain and binding of CoA for production of the final product, a fatty acyl-CoA thioester[2]. Shown below is the general mechanism for ACS proteins.
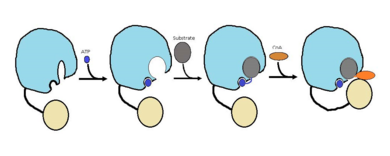
Figure 1 shows the general outline of the binding of ATP and acyl substrates to an ACSVL enzyme. This is the accepted mechanism for these types of proteins.
Background
Mycobacterium Tuberculosis is the causative agent of Tuberculosis commonly abbreviated TB. TB causes approximately 1.4 million deaths every year. The cost for treatment of patients with TB between the years 2010-2015 was approximately 16 billion dollars. TB is spread through the air, not by contact. There are two forms of TB, latent TB and TB disease.
Structural Highlights
FadD13 is an ACSVL enzyme that can accept lipids up to 26 carbons as well as being a peripheral-membrane protein[3]. Unlike other ACSVL proteins, FadD13 is soluble. There are numerous aspects of its structure that affects the way this protein functions. The is comprised of two separate motifs. The first motif is composed of residues 164-TSGTTGHPKG173-173 shown in red which binds to the phosphate group. The second motif is comprised of residues 298-VQGYALTE-305 shown in blue which binds to the adenine group[4]. Upon binding of ATP or AMP, FadD13 is activated. There are two domains of this protein, a larger N-terminal domain () shown in blue and a smaller C-terminal domain () shown in yellow. These domains are held together by a six amino acid linker () shown in black. Inside the larger N-terminal domain is a composed of six beta sheets (beta 9-14) shown in green and two alpha helices (alpha 8-9) shown in red.The hydrophobic tunnel allows large lipids/fatty acids, up to 26 carbons, to bind. The tunnel is capped by an arginine and aromatic rich shown in yellow that is involved in the peripheral binding of the enzyme to the membrane. Six key arginine residues, create a positively charged surface that is likely involved in initially recruiting FadD13 to the membrane. When these residues were replaced with hyrdrophobic alanine residues, membrane binding increased. This points to the important role of hydrophobic interactions in keeping the protein bound at the membrane[5].
Function
The FadD13 enzyme functions to activate lipids. Once the lipids are activated, they can continue on into metabolic pathways. This is done by ATP/AMP binding to the . Once ATP/AMP is bound, the long lipid chain up to 26 carbons may bind in the of the enzyme. Upon binding of the substrate, the C terminal swings up to close off the tunnel. From there CoA can bind to produce the final product, an acyl-CoA Thioester. The lipid can now move transversely throughout the membrane and throughout the rest of the cell. Below is the proposed mechanism for ACSVL proteins.
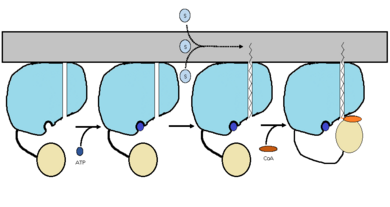
Figure 2 shows the proposed mechanism for an ACSVL protein bound to the membrane
[6]


