There are two flavors of estrogen receptors. ER - Intracellular receptor and GPER - G protein coupled receptors. ERs are transcription factors while GPER are not transcription factors. ERs are the focus of this page. Proteopedia and the provided scene editing tools were used to help create images [1] shown in this article.
Function
Like other steroid hormones estrogen effects the transcription of a large number of genes via its interactions with its intracellular receptors, estrogen receptors. Estrogen receptors are receptors that are activated by the hormone estrogen (Example of an estrogen; 17β-estradiol).
Structural highlights
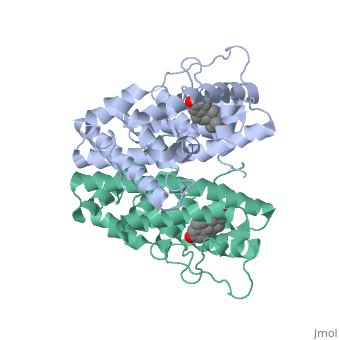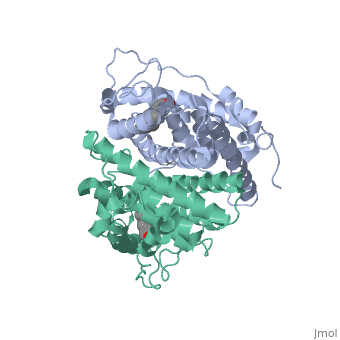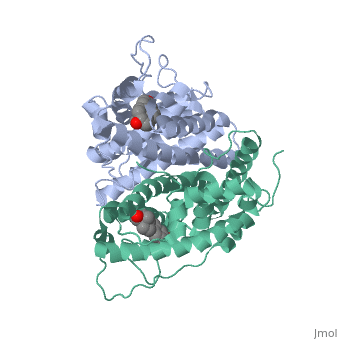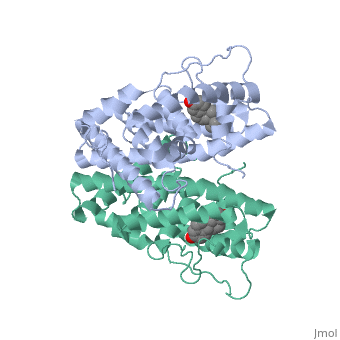
ER is a modular protein composed of a ligand binding domain, a DNA binding domain and a transactivation domain. ER is a DNA-binding transcription factor. The DNA binding domain can be clearly observed in this scene; the highlighted yellow helix in close proximety to the DNA is part of the DNA binding domain. The blue beta sheet close to the yellow DNA binding alpha helix is also part of the DNA binding domain. The transactivation domain is attached at the end of the yellow DNA binding domain, also forming an alpha helix colored in yellow. The transactivation domain activates RNA polymerase when the receptor binds to DNA. The ligand binding domain may be observed here with the following scene. The ligand ferutinine is bound by the ligand binding domain, composed of the surrounding blue colored alpha helices . Unbound ER normally exists loosly around the nucleus; this is subject to change depending on a multitude of factors including cell type, progress through cell cycle and reception of cellular signals. When estrogen enters the cell and binds ER, ER trans-locates and undergoes a conformation shift.[2] Ligand bound estrogen receptor associates more tightly with the nucleus.
Ferutinine also causes ER to form a tight loop allowing stimulation of normal growth.
DNA Protein Interaction and ER Regulation
ER is functional as a ligand-dependent transcription factor. [3] ER responds to both agonist and antagonist ligands and can associate with the nuclear matrix. Differences in the structure of the receptor are observed depending on what ligand ER has bound. Through comparisons of ER bound to agonist and antagonist ligands some structural components may be highlighted. The specific conformation of this tight loop of alpha helices and beta sheets around the ligand creates part of the activation signal that will stimulate normal growth; normal growth is stimulated by chaperon proteins recognizing the estrogen receptor ligand complex and facilitating the trans-location of the complex to the nucleus. Eventually the complex will reach specific euchromatin, at which point the chaperon protein and estrogen receptor ligand complex changes conformation so as to allow the estrogen receptor to bind the major groove at specific palindromic sequences. Estradiol is a normal ligand for ER and allows for binding in the major groove of DNA. If the ligand is an antagonist the transcription factor function of estrogen receptor becomes hindered. The conformation of ER bound to the partial agonist genistein has a loop which is not as tight around the ligand as those found on ER with a complete agonist ligand. This slight difference effects the ability of the chaperon to be able to bind the receptor ligand complex to the major groove of DNA.
This is noticeable in the size difference of the pure agonist vs partial agonist scenes. Special attention should be given to the bottom right alpha helices and beta sheets that are pushed out more in the antagonist compared to the agonist.
Tamoxifen is a drug created to bind ER and inhibit the transcription factor activity of ER. Tamoxifen is larger than the normal hormone ER binds (estradiol); for this reason added with the conformation estrogen receptor takes on, the activation loop is pushed into an inactive conformation. This blocks ER from giving the signal to grow. Antagonists are generally larger and cause estrogen receptors to be too hindered sterically to be able to bind to the major groove of DNA, inhibiting the receptor. The antagonist bound estrogen receptor is noticeably larger than the agonist bound version.
The location of the receptor bound and unbound to ligand varies amongst different cell types. In general, an antagonist ligand will cause partial accumulation in the cytoplasm of a cell. The agonist ligand causes the translocation to the nucleus described above.
A group bound GFP to ER and studied the location of GFP-ER upon binding of agonists and antagonist ligands. [4] GFP-ER activates the reporter gene in a dose-dependent manner and shows additional activation in the presence of agonist ligand 17-bestradiol. ICI 182780, a pure antagonist for ER, completely inhibited GFP-ER activation of the reporter gene.
The group found that in the absence of ligand, the unoccupied ER is loosely associated with the nucleus.
Ligand causes a biochemical transformation into a complex that associates more tightly with the nucleus.
Before ER binds its hormone the receptor is part of a complex that has many chaperones that maintain the receptor in a steroid binding configuration. Post hormone binding the receptor dissociates from its original complex and binds to hormone responsive elements in chromatin. Gene expression is then regulated by interaction of DNA bound receptors with sequence specific transcription factors and general transcription factors which are mediated by co-activators and co-repressors. The arrangement of cis regulatory elements in a specific promoter or enhancer region and the current state of DNA sequences in nucleosomes determines the system of receptor interactions. Contingent upon the interactions occurring, the result may be induction or repression of transcription.
ERα-regulated gene expression involves interactions with cointegrators (ex. p300/CBP, P/CAF) that have the capacity to modify core histone acetyl groups. 2 ER’s DNA binding domain is ordered around two zinc ions that allow the receptors to bind as homodimers to palindromic DNA sequences in such a way that each homodimer links to one half of the palindrome.
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.