From Proteopedia
proteopedia linkproteopedia link Kinesin
|
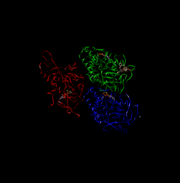 This is 4UXR, the head of kinesin.
Function
Kinesin is generally responsible for the transport of polypeptides and other large molecules throughout a single cell, such as moving the spindles during cell division. Kinesin can only move from the negative (-) end of microtubules to the positive (+) end. This means that kinesin only moves objects out towards the periphery of the cell and thus aids in the secretion of molecules or division of cells.
Disease
Kinesin are involved during mitosis, failure of kinesin to work properly can lead to trisomy and other meiosis/mitosis related diseases. [1]
Structural highlights
Heavy Chain:
It is the most conserved region amongst kinesin which consists of the head, neck, and tail. Usually contains eight core β-sheets and six major alpha helixes, most of these secondary structures are in different places in the primary sequence but line up in the tertiary structure.
Light Chain:
Not technically part of the protein of kinesin or myosin itself, but its presence is necessary for activity. It regulates conformational changes within the protein.
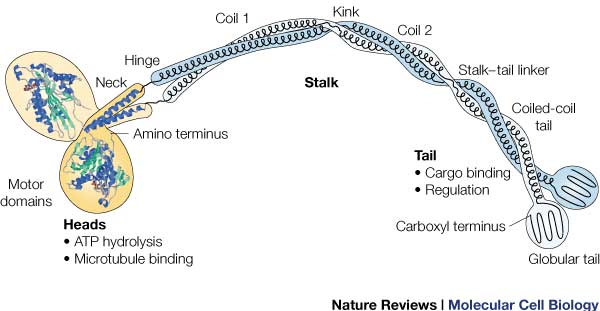
Head:
Most conserved domain amongst all kinesin, it consists mainly of alpha helixes. Its tertiary structure usually includes a large cleft from the actin binding site to the ATP binding pocket. The head has the ability to bind microtubule on one site and bind ATP at another. This section undergoes the most conformational change and is responsible for the force that causes kinesin to move along the cytoskeleton. In kinesin, binding of ATP appears to have allosteric control over the binding of kinesin to the tubules, by a twisting of a β- sheet core, showing allosteric control between the subdomains within the protein. [2]
ATPase: It generates this force using an ATPase either coupled with the myosin/kinesin or inherent within the head, the inherent ATPase uses a P-loop, which is a phosphate- binding loop used in adenylate kinase, Ras, and others. Both also create a similar environment for the γ-phosphate, which uses a conserve motif of Ser-Ser-Arg.[3]
In kinesin, the binding of ATP greatly increases the binding of kinesin to the microtubules, to allow it to perform a power stroke. The cleft uses His 93, Pro 17 and Arg 16 to hold ATP. The lack of these loops allows kinesin to react with ATP much faster.
RAS fold: conserved domain among many signaling proteins. A structural motif where a nucleotide molecule is bound to loops at one end of a β-sheet domain. This suggests that myosin and kinesin may have evolved from a common ancestor.[3]
Neck:
completely alpha-helical region between the head and the tail on the heavy chain that binds to the light chains.
In kinesin, the neck region of amino acids 330-370 generally form a coiled coil region that is long and flexible.
Tail:
Includes polypeptide binding site, varies the most since it is responsible for dictating the location within the cell that the protein is active.
|
References
Goodsell, David. "Kinesin." RCSB PDB-101. Protein Data Bank, n.d. Web. 07 Nov. 2015.
Krukau, Aliaksei, Volker Knecht, and Reinhard Lipowsky. "Allosteric Control of Kinesin's Motor Domain by Tubulin: A Molecular Dynamics Study." Phys. Chem. Chem. Phys. Physical Chemistry Chemical Physics 16.13 (2014): 6189. Royal Society of Chemistry. Web. 6 Nov. 2015.
Kull, F. Jon, Elena P. Sablin, Rebecca Lau, Robert J. Fletterick, and Ronald D. Vale. "Crystal Structure of the Kinesin Motor Domain Reveals a Structural Similarity to Myosin." Nature 380.6574 (1996): 550-55. The National Center for Biotechnology Information. Web. 6 Nov. 2015.
Lodish, Harvey F., Arnold Berk, Paul Matsudaira, Chris A. Kaiser, Monty Krieger, Matthew P. Scott, Stephen Lawrence Zipursky, and James E. Darnell. Molecular Cell Biology. New York, NY: W.H. Freemann, 2003. Print.
Paxton, Walter F., Nathan F. Bouxsein, Ian M. Henderson, Andrew Gomez, and George D. Bachand. "Dynamic Assembly of Polymer Nanotube Networks via Kinesin Powered Microtubule Filaments." Nanoscale 7.25 (2015): 10998-1004. Royal Society of Chemistry. Web. 6 Nov. 2015.
Rayment, I., W. Rypniewski, K. Schmidt-Base, R. Smith, D. Tomchick, M. Benning, D. Winkelmann, G. Wesenberg, and H. Holden. "Three-dimensional Structure of Myosin Subfragment-1: A Molecular Motor." Science 261.5117 (1993): 50-58. PubMed. Web. 6 Nov. 2015.
Rice, Sarah, Abel W. Lin, Daniel Safer, Cynthia L. Hart, Nariman Naber, Bridget O. Carragher, Shane M. Cain, Elena Pechatnikova, Elizabeth M. Wilson-Kubalek, Michael Whittaker, Edward Pate, Roger Cooke, Edwin W. Taylor, Ronald A. Milligan, and Ronald D. Vale. "A Structural Change in the Kinesin Motor Protein That Drives Motility." Nature 402.6763 (1999): 778-84. Nature. Web. 6 Nov. 2015.
"SDSU Biology 590 - Actin Myosin Crossbridge 3D Animation." SDSU Biology 590 - Actin Myosin Crossbridge 3D Animation. San Diego State University College of Sciences, 30 Sept. 97. Web. 07 Nov. 2015.
Song, Y.-H. "Structure of a Fast Kinesin: Implications for ATPase Mechanism and Interactions with Microtubules." The EMBO Journal 20.22 (2001): 6213-225. PubMed. Web. 6 Nov. 2015.
https://labs.cellbio.duke.edu/kinesin/KinesinStructure.html
- ↑ doi: https://dx.doi.org/10.2210/rcsb_pdb/mom_2005_4
- ↑ Krukau A, Knecht V, Lipowsky R. Allosteric control of kinesin's motor domain by tubulin: a molecular dynamics study. Phys Chem Chem Phys. 2014 Apr 7;16(13):6189-98. doi: 10.1039/c3cp53367k. PMID:24561904 doi:http://dx.doi.org/10.1039/c3cp53367k
- ↑ 3.0 3.1 Kull FJ, Sablin EP, Lau R, Fletterick RJ, Vale RD. Crystal structure of the kinesin motor domain reveals a structural similarity to myosin. Nature. 1996 Apr 11;380(6574):550-5. PMID:8606779 doi:http://dx.doi.org/10.1038/380550a0



