Sandbox Reserved 1124
From Proteopedia
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
|
Contents |
Introduction
Grb2 (Growth factor receptor-bound protein 2) is a connector protein that link grow factor receptors to the Ras signalling pathway. This protein implied in signal transduction pathways is essential for multiple cellular functions such as: embryonic development, cell proliferat
Structure
Grb2 is a small protein of 217 residues with a molecular size of about 25 Da and composed of three remarkable domains : a single SH2 (Src Homology 2) domain (60 to 152 pdb) flanked by two conserved SH3 domains (respectively 1 to 58 and 156 to 215 pdb). It has no catalytic domain. The central SH2 domain binds growth factor receptors (EGFR or PDGFR) or scaffold proteins. It interacts preferentially with a tyrosine phosphorylated sequence with the following motif: pY-X-N-X (X is a hydrophobic residue). The two SH3 domains bind proline-rich regions of other proteins and enable the interaction with the Sos protein (guanine nucleotide exchange factor). The N-Terminal SH3 domain plays the main role in this interaction, it binds a proline-rich motif PxxP of the Ct domain of Sos. The Ct SH3 domain improves the overall stability of the Grb2-Sos complex. Moreover, this Ct domain specifically binds to proteins with a P-X-I/L/V-D/N-R-X-X-K-P motif such as Gab1.
The SH2 domain
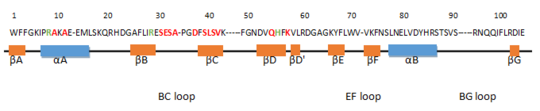
encompasses 8 beta strands (61 to 64 ; 82 to 87 ; 95 to 101 ; 104 to 109 ; 111 to 112 ; 118 to 119 ; 124 to 125 ; 149 - 150) and 2 alpha helices (67 to 74 and 128 to 134). The βB, βC and βD strands compose a three-stranded antiparallel β-sheet and the 2 α-helices are positioned on both sides. Moreover, the short parallel βA and βG strands extend the central β-sheet. There are also βD', βE and βF strands which are smaller β-sheet-like structure. [1]The amino acid in red are residues which are responsible for forming the phosphopeptide binding pocket. In green, this is residues which can bind to the negatively charged phosphorylated tyrosine residue of the binding peptide. This domain is very essential for the function of Grb2. Actually, several mutations in the SH2 domains can cause human diseases. A mutation for example of the arginin residue at position 5 of βB can abolish the phosphotyrosine dependent interactions.[2]
The N-Terminal SH3 domain
encompasse two three-stranded antiparallel β-sheets, one strand crosses the two sheets. This confers a barrel-like structure upon the domain. The first sheet contains the 3 following strands: S1 (Glu2-Ala5), S2 (Ile24-Lys26) and S6 (Ile53-Met55). The second sheet contains the strands S3 (Val27-Asn29), S4 (Trp36-Leu41) and S5 (Asp45-Ile48). The structure of this SH3 domain is stabilized by a high number of hydrophobic residues, which form the centre of the protein.
The C-terminal SH3 domain
....
Function
</StructureSection>
References
- ↑ Ogura K, Shiga T, Yokochi M, Yuzawa S, Burke TR Jr, Inagaki F. Solution structure of the Grb2 SH2 domain complexed with a high-affinity inhibitor. J Biomol NMR. 2008 Nov;42(3):197-207. doi: 10.1007/s10858-008-9272-0. Epub 2008, Oct 2. PMID:18830565 doi:http://dx.doi.org/10.1007/s10858-008-9272-0
- ↑ Kousik Kundu In Silico Prediction of Modular Domain-Peptide Interactions (2015) [1]