Sandbox Reserved 1124
From Proteopedia
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
Grb2
|
Contents |
Introduction
Grb2 (Growth factor receptor-bound protein 2) is a connector protein that link grow factor receptors to the Ras signalling pathway. This protein implied in signal transduction pathways is essential for multiple cellular functions such as: embryonic development and cell proliferation. Grb2 can be found in the cytosol, the nucleus and the plasmic membrane.
DNA/RNA
The gene which codes the Grb2 protein is located on the seventeen chromosome. It is composed of five exons, ranging from 78 to 186 bp, and four introns from 1 to 7 kb. It is transcribed into 2 mRNA arising from alternative splicing. Thus, there are two protein isoforms. The second one does not have the exon of the 3’ coding region, which is the origin of the residues from 59 to 100 in the mature Grb2. In fact, it is a deletion in the amino-terminal part of the SH2 domain. Therefore, the function is modified because this domain cannot bind the phosphorylated tyrosine.
Structure
Grb2 is a small protein of 217 residues with a molecular mass of about 25,206 Da and composed of three remarkable domains : a single SH2 (Src Homology 2) domain (60 to 152 pdb) flanked by two conserved SH3 domains (respectively 1 to 58 and 156 to 215 pdb)[1]. It has no catalytic domain. The central SH2 domain binds growth factor receptors (EGFR or PDGFR) or scaffold proteins. It interacts preferentially with a tyrosine phosphorylated sequence with the following motif: pY-X-N-X (X is a hydrophobic residue).[2] Other non-receptor tyrosine kinases have also this motif and interact with Grb2 SH2 domain, such as BCR-Abl, focal adhesion kinase, insulin receptor substrate 1 and PTPN11. The two SH3 domains bind proline-rich regions of other proteins and enable the interaction with the Sos protein (guanine nucleotide exchange factor). The N-Terminal SH3 domain plays the main role in this interaction, it binds a proline-rich motif PxxP of the Ct domain of Sos.[3] This binding region in Sos has the shape of a Polyprolin II helix. The Ct SH3 domain improves the overall stability of the Grb2-Sos complex. Moreover,this Ct domain specifically binds to proteins with a P-X-I/L/V-D/N-R-X-X-K-P motif such as Gab1. [4] The determination of the crystallographic structure enabled the observation of the junction between the SH3 and SH2 domains. It allows the two adjacent faces of the SH3 domains to be closer. This association is strenghtened by Van der Waals interaction. Nevertheless, the proline-rich peptides can still bind to SH3 domains.
The SH2 domain
encompasses 8 beta strands ( ; ; ; ; ; ; ; ) and 2 alpha helices ( and )[5]. The βB, βC and βD strands compose a three-stranded antiparallel β-sheet and the 2 α-helices are positioned on both sides. Moreover, the short parallel βA and βG strands extend the central β-sheet. There are also βD', βE and βF strands which are smaller β-sheet-like structure. [6]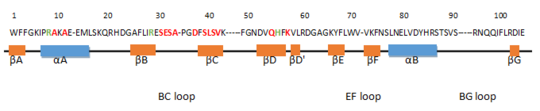
The amino acid in red are residues which are responsible for forming the phosphopeptide binding pocket. In green, this is residues which can bind to the negatively charged phosphorylated tyrosine residue of the binding peptide. This domain is very essential for the function of Grb2. Actually, several mutations in the SH2 domains can cause human diseases. A mutation for example of the arginin residue at position 5 of βB can cancel the phosphotyrosine dependent interactions.[7]
The SH2 domain of Grb2 enables the interaction with receptors, scaffold proteins, tyrosine kinases but also with other adaptor proteins. Indeed, Shc is an intermediate between some receptors and Grb2.
The N-Terminal SH3 domain
encompasse two three-stranded antiparallel β-sheets, one strand crosses the two sheets. This confers a barrel-like structure upon the domain. The first sheet contains the 3 following strands: S1 (Glu2-Ala5), S2 (Ile24-Lys26) and S6 (Ile53-Met55). The second sheet contains the strands S3 (Val27-Asn29), S4 (Trp36-Leu41) and S5 (Asp45-Ile48). The structure of this SH3 domain is stabilized by a high number of hydrophobic residues, which form the centre of the protein.[3]
The C-terminal SH3 domain
....
Function
MAP kinases pathway
In the cytosol, Grb2 is bound to the guanine nucleotide exchange factor SOS-1 via its SH3 domain. Then, this complexe is recruited to the plasmic membrane to be close the Ras protein. To help Grb2 binds the phosphorylated tyrosines of the EGFR via its two SH2 domains. The Ras protein is a small GTPase which is in an inactive state when it is bound to GDP. However, the exchange of GDP for GTP actives it allowing the bond and the activation of Raf 1, a serine/threonine protein kinase. A cascade of kinase phosphorylation is then initiated. Indeed, Raf 1 phosphorylates MEK1 or MEK 2 which in turn phosphorylate ERK1 or ERK2. Finally, these MAP kinases allow the translocation of transcription factors to the nucleus and their phosphorylation. Such as STAT 1 or Elk-1. Moreover, the SH2 domain recognizes the C-terminal domain of FAK (Focal Adhesion Kinase) called the focal adhesion targeting region (FAT) when its tyrosine 925 on the first helix is phosphorylated. But, this tyrosine contains the pY925 motif and is not a β-turn structure. Thus, an adaptation of this tyrosine on the SH2 domain is possible. To conclude, the interaction between Grb2 and FAK leads to the activation of the Ras-MAPK pathway.
PI3K/AKT pathway
In epithelial cells, an adaptator called Gab1 binds the carboxyl terminal SH3 domain of Grb2. This causes the activation of PI3K/AKT pathway.
T cells
When TCR are stimulated, the membrane protein called p36-38 is phosphorylated in the T cells. Then, the phosphorylated tyrosines bind the SH2 domains of Grb2 whereas the SH3 domains are bound to Vav proteins. These interactions allow the T cell proliferation, the calcium flux in these cells and the MAP kinase activation.
</StructureSection>
References
- ↑ Gagani Athauda, Donald P Bottaro Atlas of Genetics and Cytogenetics in Oncology and Haematology (2007)[1]
- ↑ Wikipedia Grb2 [2]
- ↑ 3.0 3.1 doi: https://dx.doi.org/10.1038/nsb1294-898
- ↑ doi: https://dx.doi.org/10.1016/S0960-9822(02)01038-2
- ↑ UniProtKB P62993 Human
- ↑ Ogura K, Shiga T, Yokochi M, Yuzawa S, Burke TR Jr, Inagaki F. Solution structure of the Grb2 SH2 domain complexed with a high-affinity inhibitor. J Biomol NMR. 2008 Nov;42(3):197-207. doi: 10.1007/s10858-008-9272-0. Epub 2008, Oct 2. PMID:18830565 doi:http://dx.doi.org/10.1007/s10858-008-9272-0
- ↑ 7.0 7.1 Kousik Kundu In Silico Prediction of Modular Domain-Peptide Interactions (2015) [3]
