From Proteopedia
proteopedia linkproteopedia link
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159.
|
To get started:
- Click the edit this page tab at the top. Save the page after each step, then edit it again.
- Click the 3D button (when editing, above the wikitext box) to insert Jmol.
- show the Scene authoring tools, create a molecular scene, and save it. Copy the green link into the page.
- Add a description of your scene. Use the buttons above the wikitext box for bold, italics, links, headlines, etc.
More help: Help:Editing
|
Plasmodium falciparum Atg8 in complex with Plasmodium falciparum Atg3 peptide
| 4eoy is the PDB code corresponding to a proteic complex composed of two structures : Atg8 and Atg3. These proteins belong to an organism named Plasmodium falciparum, a parasite which is responsible for malaria. They play a part in autophagy, a process which consists in recycling a part of the cytoplasm in order to make the cell go from the sporozoite (growing) state to the erythrocytic infective state.
Function
Cellular effects
The autophagy related proteins (Atg) are proteins which purpose is to help the parasite to survive and develop. Atg8 and Atg3 are both involved in the autophagy process, by creating an autophagosome and then making it grow. The autophagosome can be compared to a vacuole and absorbs the components of the cytoplasm (in a non specific way). When the growing phase (sporozoite) of the cell is over,in the case of Plasmodium falciparum, the autophagy is launched by the cell in order to become erythrocytic-infective. This is the time when the cell starts its viral activity. Lysosomes then merge with the autophagosme to form the autolysosome. Once this process is done, the autolysosome can complete its purpose : recycle the its content. In fact, all the molecules inside are degradated and their components (like aminoacids) are reused in order to synthetise viral proteins.
Molecular effects
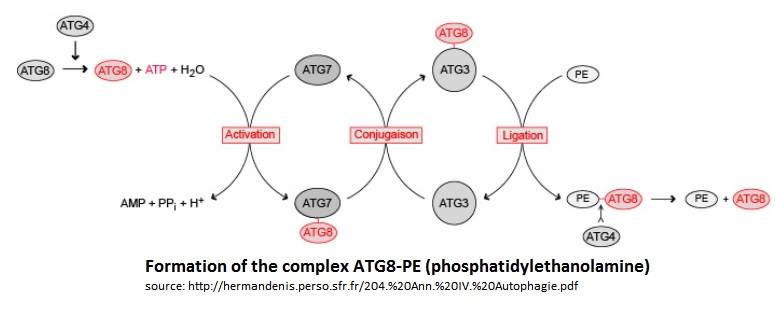
In the Atg8 system, there are several steps until Atg8 is activated. Firstly, the cysteine protease Atg4 cleaves the last residue of the C-terminal domain of Atg8 by consuming 1 molecule of ATP and H20. Then, the Atg7 (E1-like enzyme) activates the C-terminal glycine exposed and therefore forms an intermediate Atg8-Atg7 by creating a thioester bond. Atg3 (E2-like enzyme) then replaces Atg7 in the complex to form an Atg8-Atg3 thioester intermediate. Phosphatidylethanolamine (PE) then links to the N-terminal domain of Atg8. This final complex (Atg8-Atg3-PE) serves as an intermediate for membrane tethering and hemifusion of the autophagosome. This complex is primordial for the elongation of the autophagosome. Notice that Atg8-Atg3-PE is the active complex, but Atg8-PE can also be sufficient to do this task (like this is showed on the scheme below).
Disease
Malaria is an infectious and lethal disease transmitted by Anopheles mosquitos, leading to the apparition of fever, vomiting, fatigue or even coma. If this disease is not well treated, the symptoms could return even months after and can infect humans as well as other animals.
The parasite, a plasmodium type (Plasmodium falciparum for humans), introduced by the female mosquito, will infect the hepatic cells then the victim’s blood in order to destroy the red blood corpuscule.
Relevance
Malaria is a strikening disease killing more than millions of people every years, primarily children. This is one of the most killing disease on earth at this time. That's why it is essential to focus the medical research on malaria's treatment.
Recent studies (2012) have shown a start point for drug researches. In fact 1,2,3-tihydroxybenzene have shown to prevent protein-protein interaction between Atg8 and Atg3, inhibiting autophagosome's elongation and leading to the no-infectiveness of the parasite.
Structural highlights
Atg3 protein
Atg3 is a protein composed of 314 aminoacids with an alpha/beta-fold with a core region topologically similar to E2 enzymes . The core region has two regions:
→ the first region has 80 residues and has a random coil structure in solution, this region is responsible for the Atg7 interaction which is an E1-like enzyme.
→ The second is an alpha-helical structure which protrudes from the core region. This alpha-helical structure is responsible for binding Atg8.
Moreover some researches indicates that the catalytic cysteine of Atg3 is a possible binding site for a phosphate of phosphatidylethanolamine.
Atg8 protein
Atg8 is a protein of 117 aminoacids with a molecular wieght of 13,6kDa. This molecule is composed of 5- stranded β-sheet. Those β-sheet are enclosed by two α-helices on each sides. This conformation leaves accessible a conserved GABARAP domain, this protein has been originally identified as a binding partner of a GABAA receptor subunit. GABARAP. Even if the sequences between Atg8 and ubiquitin are not similars, the crystal structure reveals a conserved ubiquitine-like fold. Atg8 belongs to the ATG family but it differs from the other members of the family because the α2 helix-terminating proline 26 was substituted by a lysine.
Atg8/Atg3 Complex
Atg3 and Atg8 interact through a thioester bond between the Cys-288 of Atg3 and the C-terminal Glycine of Atg8.
|
References
1. Hain AU, Weltzer RR, Hammond H, Jayabalasingham B, Dinglasan RR, Graham DR, Colquhoun DR, Coppens I, Bosch J
Structural characterization and inhibition of the plasmodium atg8-atg3 interaction.
J.Struct.Biol. (2012) 180 p.551
2. Machiko Sakoh-Nakatogawa, Kazuaki Matoba, Eri Asai, Hiromi Kirisako, Junko Ishii, Nobuo N Noda, Fuyuhiko Inagaki, Hitoshi Nakatogawa & Yoshinori Ohsumi
Atg12–Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site
Nature Structural & Molecular Biology 20, 433-439 (2013) doi :10.1083/nsmb.2527
3. Oliver H. Weiergräber, Jeannine Mohrlüder and Dieter Willbold
Atg8 Family Proteins — Autophagy and Beyond, DOI: 10.5772/55647
Biochemistry, Genetics and Molecular Biology » "Autophagy - A Double-Edged Sword - Cell Survival or Death?", book edited by Yannick Bailly, ISBN 978-953-51-1062-0, Published: April 17, 2013 under CC BY 3.0 license.
4. Herman Denis, http://hermandenis.perso.sfr.fr/204.%20Ann.%20IV.%20Autophagie.pdf
5. Patrice Codogno
Les gènes ATG et la macro-autophagie
Med Sci (Paris). 2004 August; 20(8-9): 734–736.
Published online 2004 August 15. doi: 10.1051/medsci/2004208-9734
6. Hain AU, Bartee D, Sanders NG, Miller AS, Sullivan DJ, Levitskaya J, Meyers CF, Bosch J.
Identification of an Atg8-Atg3 protein-protein interaction inhibitor from the medicines for Malaria Venture Malaria Box active in blood and liver stage Plasmodium falciparum parasites.
J Med Chem. 2014 Jun 12;57(11):4521-31. doi: 10.1021/jm401675a. Epub 2014 May 19
7. Masaya Yamaguchi, Nobuo N. Noda, Hitoshi Nakatogawa, Hiroyuki Kumeta, Yoshinori Ohsumi and Fuyuhiko Inagaki
Autophagy-related Protein 8 (Atg8) Family Interacting Motif in Atg3 Mediates the Atg3-Atg8 Interaction and Is Crucial for the Cytoplasm-to-Vacuole Targeting Pathway.
The Journal of Biological Chemistry vol.28. 2010, September 17. doi:10.1074/jbc.M110.11367
8. Hain AU1, Weltzer RR, Hammond H, Jayabalasingham B, Dinglasan RR, Graham DR, Colquhoun DR, Coppens I, Bosch J.
Structural characterization and inhibition of the Plasmodium Atg8-Atg3 interaction. J Struct Biol. 2012 Dec;180(3):551-62. doi: 10.1016/j.jsb.2012.09.001.
Epub 2012 Sep 13.PMID:22982544
9. Machiko Sakoh-Nakatogawa, Kazuaki Matoba, Eri Asai, Hiromi Kirisako, Junko Ishii, Nobuo N Noda, Fuyuhiko Inagaki, Hitoshi Nakatogawa & Yoshinori Ohsumi.
Atg12–Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site.
Nature Structural & Molecular Biology. 2013 February. doi:10.1038/nsmb.2527

