Dicer
From Proteopedia
Contents |
Dicer
| |||||||||||
References
1. Bernstein, E, Caudy, A, Hammond, S, and Hannon, G. (2001) Role for a Bidentate ribonuclease in the initial step of RNA interference. Cold Spring Harbor Laboratory. Nature, Vol 409, pgs. 363-367
2. MacRae, I. (2006) Structural Basis for Double-Stranded RNA processing by Dicer. Science, Vol 311, pgs. 195-198
3. Hammond, S. (2005) Dicing and Slicing: The Core machinery of the RNA interference pathway. University of North Carolina. Federation of European Biochemical Societies, Letters 579, pgs. 5822-5829
4. Khaiwesh, B, Asif Arif, M, Seumel, G, Ossowski, S, Weigel, D, Reski, R, and Frank, W. (2010) Transcriptional control of gene expression by microRNAs. Cell, Vol 140, pgs. 111-122
5. Lau, P, Potter, C, Carragher, B, MacRae, I. (2009) Structure of the Human dicer-TRBP complex by Electron Microscopy. Cell, October 14, 2009, pgs. 1326-1332.
- ↑ Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001 Jan 18;409(6818):363-6. PMID:11201747 doi:http://dx.doi.org/10.1038/35053110
Introduction
Dicer is a type of Ribonuclease that processes potentially harmful double-stranded RNA (dsRNA) into microRNA and small-interfering RNA (siRNA) to be used in the process of RNA interference. Dicer is commonly utilized by cells in order to prevent the assimilation of viral DNA into the cells’ genome. The viral DNA is butchered into smaller segments that are each about 21 nucleotides long; the cut take places at the 5’ phosphate and the 3’ hydroxyl, and usually includes a 2 nucleotide overhang. There is a single processing center in HS Dicer implying that there are two catalytic sites which help form products with the 2 3' overhang. These newly formed segments attach themselves to single stranded mRNA which ultimately leads to mRNA degradation by the cell and translational suppression. The dicer enzyme in humans contains three domains: the , , and the .[1] There are three classes of RNase III proteins which are divided into categories called Escherichia coli RNase III, , and Dicer which are given the numbers one, two, and three respectively. The Escherichia coli RNase III class has one domain while the Drosha and dicer have two domains each. There is no evidence of the first class of enzymes in mammals.
Dicer, or endoribonuclease Dicer, was discovered/named in 2001 by Emily Bernstein. She was a graduate student in Greg Hannon's lab at the Cold Spring Harbor Laboratory in New York. She was trying to discover the enzyme that was responsible for removing small RNA fragments from double-stranded RNA. The dicer enzyme was found by isolating it from the RISC complex in the RNAi mechanism. It was known that RISC was not responsible for chopping up these small RNA fragments, so this complex was isolated from the system to locate the enzyme that was the source for these RNA fragments.[2]
Dicer is a member of the RNase III family, is known as drosophilia CG4792 and it is found in multiple organisms.(2,3) The discovery of Dicer was important for understanding the regulation of gene expression and the epigenetic silencing of genes by miRNA. The human endoribonuclease Dicer is 219 kDa, which is larger than many other organisms' Dicer enzymes. This is due to Humans having different domains present, and in many cases, more domains.(3)
Structure
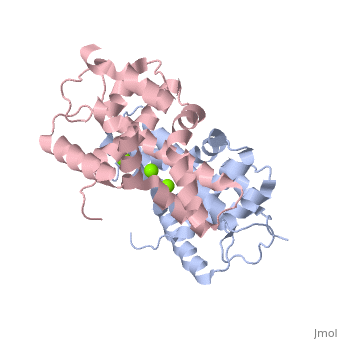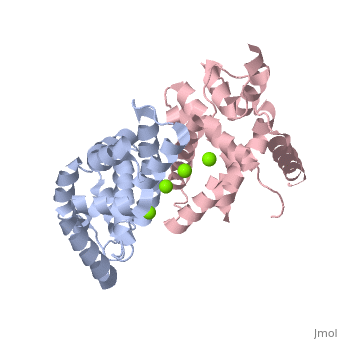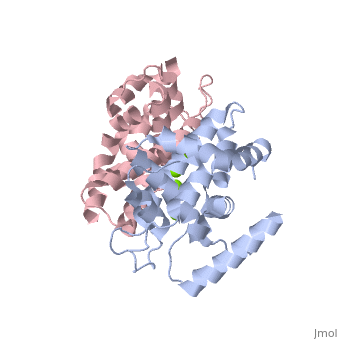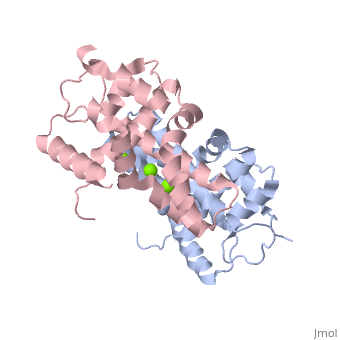
Human Dicer (hDicer) is a . Each chain is a large multidomain enzyme whose C-terminal half includes a PAZ domain, a pair of tandem RNase III domains, and a double-stranded RNA-binding domain.[3] There are four ions that bind to the hDicer RNase IIIb homodimer. There are oxygen ligands bonded to each Magnesium, which create an geometry on each Magnesium. The amino acids present on the oxygen ligands are Glutamic Acid and Aspartic Acid.
Pathology
Mutations involving the dicer protein have been linked to the development of diseases in humans. Conditions such as pleuropulmonary blastoma[4], goiter multinodular[5], and rhabdomyosarcoma[6] are related to dicer malfunction. Pleuropulmonary blastoma, goiter multinodular, cystic nephroma, and Sertoli-Leydig cell tumors are due a mutation in the Dicer1 gene given the name Dicer1 Syndrome. Dicer1 Syndrome is an inherited disorder that causes the risk of malignant tumors and benign tumors to increase. This occurs because short Dicer proteins are formed that cannot help in the production of miRNA, which can cause cells to grow into tumors. The risk of tumors is mainly increased in the lungs, kidneys, ovaries, and thyroid. Dicer1 Syndrome is transferred in an autosomal dominant pattern. The top treatment is surgery to remove the tumor. Dicer is known to be a direct cause of macular degeneration. The abscence of Dicer in retinal pigment epithelium causes the eye to break down into macular degeneration. It is hypothesized that Dicer has a specific role in maintaining this retinal health.(5)
References
- ↑ Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006 Jan 13;311(5758):195-8. PMID:16410517 doi:311/5758/195
- ↑ Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001 Jan 18;409(6818):363-6. PMID:11201747 doi:http://dx.doi.org/10.1038/35053110
- ↑ Takeshita D, Zenno S, Lee WC, Nagata K, Saigo K, Tanokura M. Homodimeric structure and double-stranded RNA cleavage activity of the C-terminal RNase III domain of human dicer. J Mol Biol. 2007 Nov 16;374(1):106-20. Epub 2007 Sep 8. PMID:17920623 doi:10.1016/j.jmb.2007.08.069
- ↑ Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, Jarzembowski JA, Wikenheiser-Brokamp KA, Suarez BK, Whelan AJ, Williams G, Bracamontes D, Messinger Y, Goodfellow PJ. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009 Aug 21;325(5943):965. doi: 10.1126/science.1174334. Epub 2009 Jun, 25. PMID:19556464 doi:10.1126/science.1174334
- ↑ Rio Frio T, Bahubeshi A, Kanellopoulou C, Hamel N, Niedziela M, Sabbaghian N, Pouchet C, Gilbert L, O'Brien PK, Serfas K, Broderick P, Houlston RS, Lesueur F, Bonora E, Muljo S, Schimke RN, Bouron-Dal Soglio D, Arseneau J, Schultz KA, Priest JR, Nguyen VH, Harach HR, Livingston DM, Foulkes WD, Tischkowitz M. DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. JAMA. 2011 Jan 5;305(1):68-77. doi: 10.1001/jama.2010.1910. PMID:21205968 doi:10.1001/jama.2010.1910
- ↑ Foulkes WD, Bahubeshi A, Hamel N, Pasini B, Asioli S, Baynam G, Choong CS, Charles A, Frieder RP, Dishop MK, Graf N, Ekim M, Bouron-Dal Soglio D, Arseneau J, Young RH, Sabbaghian N, Srivastava A, Tischkowitz MD, Priest JR. Extending the phenotypes associated with DICER1 mutations. Hum Mutat. 2011 Dec;32(12):1381-4. doi: 10.1002/humu.21600. Epub 2011 Oct 11. PMID:21882293 doi:10.1002/humu.21600
Proteopedia Page Contributors and Editors (what is this?)
Justin Woodard, Sam Hayes, Michal Harel, Ann Taylor, Wally Novak, Alexander Berchansky