Prinivil/Sandbox 1
From Proteopedia
Contents |
[1]Lisinopril
|
TEMP=Green links for structure TEMP=Needs Editing
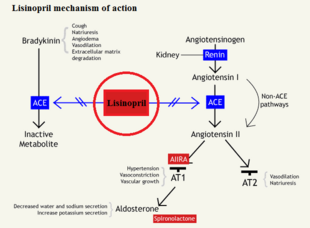
Lisinopril (Commercially known as Prinivil) functions as a competitive inhibitor of the angiotensin converting enzyme (ACE). ACE cleaves specific residues of an inactive angiotensin 1 near the C domain (domain closer to the C-terminus) to form a potent vasopressor octapeptide known as angiotensin 2; however the specific substrate binding and catalysis is not fully understood. ACE is found as a type 1 membrane bound dipeptidyl carboxypeptidase that regulates blood pressure and steady state equilibrium of ions within the blood.
Drug Function
Bradykinin is a common vasodilator that, when bound to ACE, will allow ACE the cleave angiotensin I to angiotensin II, but when bound to lisinopril, bradykinin levels rise in the blood stream causing a decrease in blood pressure and increased vasodilation. As seen in Figure 1, bradykinin will bind to the ACE, which allows for the conversion of angiotensin I to angiotensin II by the ACE/bradykinin complex. By blocking the active site of ACE, an inactive angiotensin 1 cannot be cleaved, which would block the cascade of events triggered by angiotensin 2 binding to type 1 AT2 receptor to create vasoconstriction.
Structure
Located on vascular epithelial cells, the drug interaction takes place on the surface of the cell within an artery/vein.[2] Lisinopril (prinivil) acts upon the membrane protein by forming tight and nonspecific contacts with the conserved residues in the S2’, S1’, and S1 positions of the [3]. The ligand is stabilized by 3 residues (2 histidines (RES #) and 1 glutamic acid (RES #)) We need to add the specific residue bindings here at some point interacting through hydrogen bonding along with an ionic binding of a Zinc atom and the carboxylate group which configures the molecule in 3D space.
Mechanism
Effect in Humans
TEMP
JSmol in Proteopedia [4] or to the article describing Jmol [5] to the re
</StructureSection>
References
- ↑ Canner, D. Lisinopril http://proteopedia.org/wiki/index.php/prinivil (accessed Nov 10, 2016).
- ↑ Natesh R, Schwager SL, Sturrock ED, Acharya KR. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature. 2003 Jan 30;421(6922):551-4. Epub 2003 Jan 19. PMID:12540854 doi:http://dx.doi.org/10.1038/nature01370
- ↑ Akif M, Georgiadis D, Mahajan A, Dive V, Sturrock ED, Isaac RE, Acharya KR. High-resolution crystal structures of Drosophila melanogaster angiotensin-converting enzyme in complex with novel inhibitors and antihypertensive drugs. J Mol Biol. 2010 Jul 16;400(3):502-17. Epub 2010 May 19. PMID:20488190 doi:10.1016/j.jmb.2010.05.024
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644