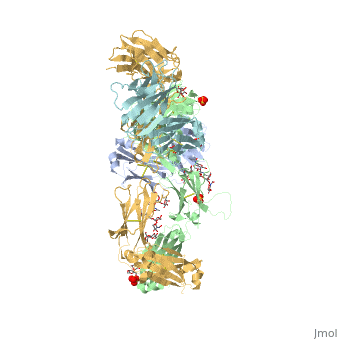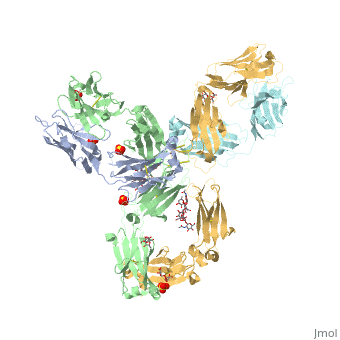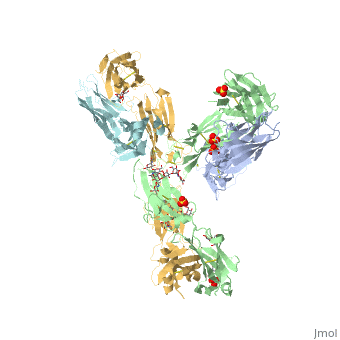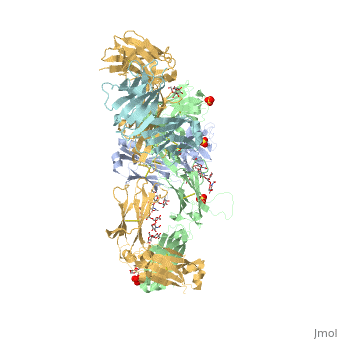Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Structure and Function
Pembrolizumab, or Keytruda, is an immunoglobulin G4 (IgG4)-kappa humanized monoclonal antibody against the programmed cell death-1 (PD-1) receptor. It contains an Fv fragment (PemFv) and a Fab fragment (PemFab). The Fv fragment is the variable region of the molecule where binding occurs whereas the Fab fragment constitutes the entire molecule. Pembrolizumab is a very compact molecule with an asymmetrical Y-shape. The short compact hinge region inflicts constraints on the molecule that creates the abnormal crystallizable tail region (Fc domain) compared to other immunoglobulin G (IgG) proteins. The Fc domain is at both CH</2> domains on each chain and one of them is distinctively rotated 120° compared to other similar structures, making the glycan chain more solvent accessible and facing the solvent. IgG4s have a unique function where they form dynamic bispecific antibodies by exchanging half-molecules (one heavy chain/light chain pair) among themselves, called Fab-arm exchange. This makes the molecule particularly unstable and unpredictable as a treatment, but is conquered by introducing the serine-to-proline mutation at , which prevents Fab-arm exchange and stabilizes the molecule [3].
Pembrolizumab/PD-1 Interaction
In order for Pembrolizumab to block PD-1, Pembrolizumab forms a large, flat paratope (antigen-binding site) that can sustain PD-1’s large epitope (where antibody attaches on antigen). The induced interaction between Pembrolizumab and PD-1 gives rise to a surface conformational change on PD-1. The new structure of PD-1 becomes a very shallow, “crescent”-like shape, in contrast to it’s flat conformation when bound to PD-L1 [4].
PemFv/PD-1 Interaction
The Fv fragment of Pembrolizumab can form a complex with the extracellular domain (ECD) of PD-1. Both PemFv and PD-1ECD contain interchain disulfide bonds. PemFv interacts predominantly in the major groove of PD-1, which is formed on one surface by the CC’FG antiparallel β−sheet and the BC, C’D, and FG loops. There are 15 direct hydrogen bonds between the residues, 15 water-mediated hydrogen bonds, 2 salt bridges, and many hydrophobic interactions. A very large solvent-accessible surface area of 1,137Å2 is buried on PD-1ECD due to the convoluted interaction. There are a total of 26 PD-1ECD residues involved in the interaction with PemFv, with residues in loop C’D (Pro84 to Gly90) and strand C’ (Gln75 to Lys 78) playing a major role. These key components of PD-1 mainly form interactions through salt bridges and hydrogen bonds with CRD-L3, CDR-H1, CDR-H2, CDR-H3 of Pembrolizumab. It is believed that the sugar chains of PD-1 have no physical contact with Pembrolizumab due to the N-linked glycosylated residues (Asn49, Asn58, Asn74, and Asn116) being located away from the interface [4].
PD-L1/PD-1 Interaction
The complex formed when protein-derived ligand, PD-L1, interacts with the inhibitory receptor, PD-1, suppresses immune responses against autoantigens and helps in peripheral immune tolerance. However, when tumors over express PD-L1, the interaction with PD-1 inhibits T-lymphocyte proliferation, release of cytokines, and cytotoxicity, exhausting tumor-specific T-cells. There are a total of 12 PD-1ECD residues that are involved in forming the complex with the N-terminal half of PD-L1ECD (PD-L1ECD-N). Nine hydrogen bonds, 3 water-mediated hydrogen bonds, 2 salt bridges, and numerous hydrophobic interactions make up the PD-1ECD/PD-L1ECD-N interaction.The CC’FG sheet within both proteins is the main interaction point. A hydrophobic surface patch is formed when the PD-1ECD is in complex with PD-L1ECD-N. The PD-1ECD residues involved include Val64, Tyr68, Ile126, Leu128, Ala132 and Ile134. Numerous hydrophilic amino acids that encircle PD-L1ECD-N form salt bridges and hydrogen bonds with Asn66, Tyr68, Gln75, Thr76, Asp77, Lys78, Ala132 and Glu136 of PD-1ECD [4].
Disease in Humans - Cancer
T-cells are a major component of the immune response in the human body. They have the ability to recognize cancer-related antigens as non-self and eliminate those cells [5]. PD-L1 and PD-L2 are ligands expressed by some tumors and inhibit t-cell function when bound to PD-1, which is located on the surface of antigen-specific t-cells [6]. When PD-L1 is ligated to PD-1 an adaptive immune response occurs, and this allows cancer cells to bypass the immune surveillance and grow uncontrollably. Pembrolizumab is an FDA-approved treatment that works as a PD-1 pathway inhibitor to fight numerous forms of cancer, such at metastatic melanoma and non-small cell lung cancer. As an inhibitor, Pembrolizumab targets the cell death of PD-1 and blocks the immune checkpoint pathway. Pembrolizumab has a very high affinity to PD-1, allowing it to block the interaction between PD-1 with PD-L1 and PD-L2 very efficiently. It antagonizes the interaction between PD-1 and its known ligands, and re-activates anti-tumor immunity [3]. The PD-1/PD-L1 interaction inhibits t-lymphocyte proliferation, releases cytokines and cytotoxicity, and exhausts tumor-specific t-cells. The inhibition of this pathway reverses the exhausted t-cell phenotype and normalizes the anti-tumor response. One downside of Pembrolizumab is that it may cause inflammatory side effects [4].
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.