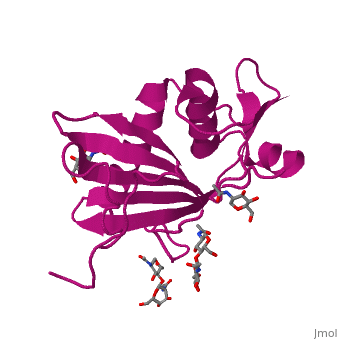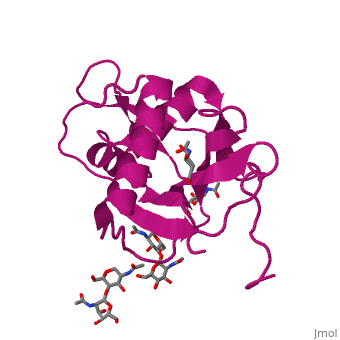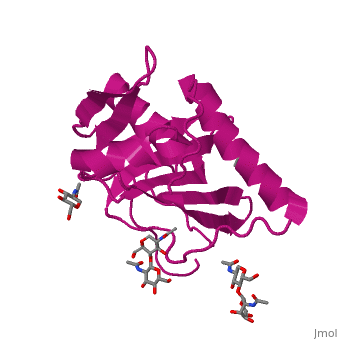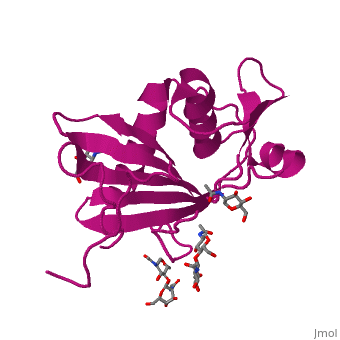Importance
Lassa virus (LASV), an Old World (OW) arenavirus, is a notorious disease-causing agent primarily in West Africa that is able to spread from rodents to humans. This deadly pathogen causes severe viral hemorrhagic fevers and significant mortality. So far, there are no available vaccines for LASV, and only one successful vaccine against another virus found in the Arenaviridae family: Junin virus[1]. Structural data at atomic resolution for viral proteins are laying the foundation for better understanding both the biology behind viral proteins and ways to combat against them. Determining the structure of the complete trimeric glycoprotein complex (GPC), composed of GP1, GP2, and SSP (stable signal peptide), will pave the path towards a future discovery of novel antiviral drugs. This is the first representative structure for OW arenaviruses. This structure reveals the overall architecture of GP1 domains from OW arenaviruses and important information relating to the mechanisms for pH switching and the binding of LASV to LAMP1 (Lysosome-associated membrane glycoprotein), a recently identified host receptor that is critical for successful infection. In addition, structural analysis suggests two novel immune evasion mechanisms that LASV may utilize to escape antibody-based immune response.
Function
GP1 (Glycoprotein 1) is the receptor binding domain of LASV that mediates receptor recognition. Research thus far indicates that GP1 from LASV may undergo irreversible conformational changes that could serve as an immunological decoy mechanism. Arenaviruses utilize various cell surface proteins as their cellular receptors for recognizing and attaching to target cells. New World (NW) arenaviruses that belong to clades A and B use transferrin receptor 1 (TfR1)[2][3], whereas OW arenaviruses, as well as clade C NW arenaviruses, use α-dystroglycan (α-DG) [4][5][6]. A trimeric class 1 viral glycoprotein complex (the spike complex) recognizes the cellular receptors and mediates membrane fusion upon exposure to low pH at the lysosome [7]. The spike complex is expressed as a glycoprotein precursor that is cleaved into three segments by a signal peptidase and SKI-1/S1P protease[8]. The functional spike complex consists of GP1, a membrane-anchored fusion protein (GP2), and a unique structured SSP [9].
Structural Highlights
GP1 of LASV is a single chain structure with attached glycans. The overall architecture of GP1 features a central β-sheet and two distinct halves: a glycosylated half containing the receptor-binding site that is made mostly by the central β-sheet and surrounding loops and a half that contains mostly helices and most likely faces the trimer axis[10], as determined in the Diskin laboratory at the Weizmann Institute of Science. The method used to determine this structure was X-ray diffraction
LAMP1 Binding Site
The primary cellular receptor of LASV is α-dystroglycan (α-DG)[11][12], which is recognized by a trimeric class 1 viral GPC (spike complex) on the viral surface[13][14]. Following successful attachment to α-DG on cells, LASV is internalized via macropinocytosis[15], and the GPC facilitates membrane fusion at the acidic environment of a late endosomal compartment[13]. Recent studies have shown that successful infection by LASV requires it to switch in a pH-dependent manner from α-DG to LAMP1[16][17]. Binding of the endosomal compartment triggers the spikes.
Histidine Triad
Included in this structure is a unique that is highly conserved among OW arenaviruses. Located on the β-sheet face of GP1, the histidine triad is a structural element that directly interacts with LAMP1 and helps stabilize a LAMP1-"compatible" conformation by providing a molecular mechanism for the pH-dependent receptor switching[10]. The is critical in forming a for LAMP1[16][18].
Resources
For more information on this protein structure visit the following sites: FirstGlance, OCA, PDBe, RCSB, PDBsum, ProSAT