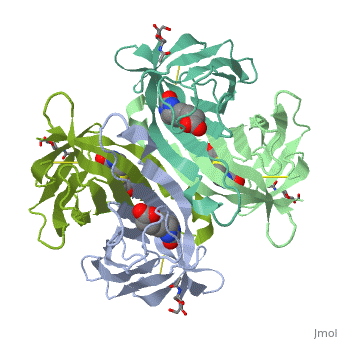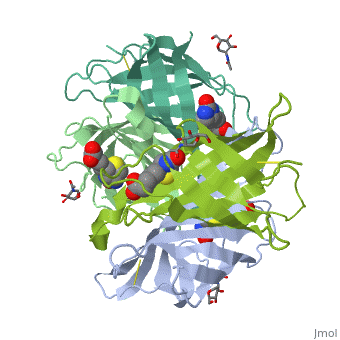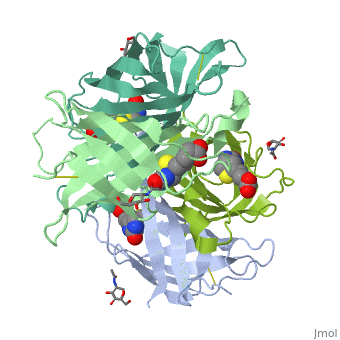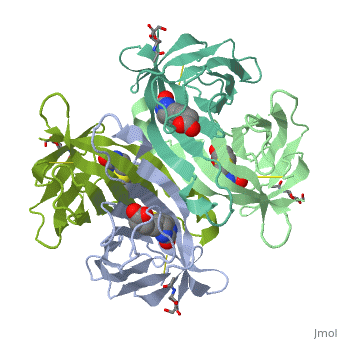2avi
From Proteopedia
| |||||||
| , resolution 3.0Å | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
THREE-DIMENSIONAL STRUCTURES OF AVIDIN AND THE AVIDIN-BIOTIN COMPLEX
Overview
The crystal structures of a deglycosylated form of the egg-white glycoprotein avidin and of its complex with biotin have been determined to 2.6 and 3.0 A, respectively. The structures reveal the amino acid residues critical for stabilization of the tetrameric assembly and for the exceptionally tight binding of biotin. Each monomer is an eight-stranded antiparallel beta-barrel, remarkably similar to that of the genetically distinct bacterial analog streptavidin. As in streptavidin, binding of biotin involves a highly stabilized network of polar and hydrophobic interactions. There are, however, some differences. The presence of additional hydrophobic and hydrophilic groups in the binding site of avidin (which are missing in streptavidin) may account for its higher affinity constant. Two amino acid substitutions are proposed to be responsible for its susceptibility to denaturation relative to streptavidin. Unexpectedly, a residual N-acetylglucosamine moiety was detected in the deglycosylated avidin monomer by difference Fourier synthesis.
About this Structure
2AVI is a Single protein structure of sequence from Gallus gallus. Full crystallographic information is available from OCA.
Reference
Three-dimensional structures of avidin and the avidin-biotin complex., Livnah O, Bayer EA, Wilchek M, Sussman JL, Proc Natl Acad Sci U S A. 1993 Jun 1;90(11):5076-80. PMID:8506353
Page seeded by OCA on Mon Mar 31 01:57:49 2008