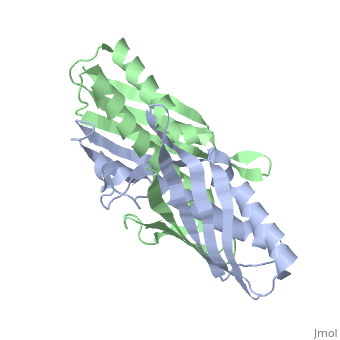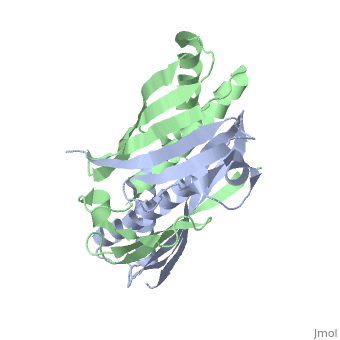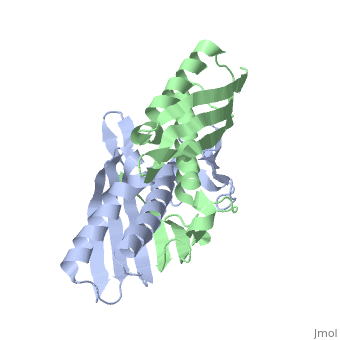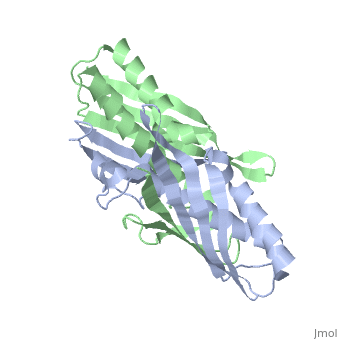
Structural highlights
Function
[OSMC_ECOLI] Preferentially metabolizes organic hydroperoxides over inorganic hydrogen peroxide.[1]
Evolutionary Conservation
Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf.
Publication Abstract from PubMed
The crystal structure of an osmotically inducible protein (OsmC) from Escherichia coli has been determined at 2.4 A resolution. OsmC is a representative protein of the OsmC sequence family, which is composed of three sequence subfamilies. The structure of OsmC provides a view of a salt-shock-induced protein. Two identical monomers form a cylindrically shaped dimer in which six helices are located on the inside and two six-stranded beta-sheets wrap around these helices. Structural comparison suggests that the OsmC sequence family has a peroxiredoxin function and has a unique structure compared with other peroxiredoxin families. A detailed analysis of structures and sequence comparisons in the OsmC sequence family revealed that each subfamily has unique motifs. In addition, the molecular function of the OsmC sequence family is discussed based on structural comparisons among the subfamily members.
Structure of OsmC from Escherichia coli: a salt-shock-induced protein.,Shin DH, Choi IG, Busso D, Jancarik J, Yokota H, Kim R, Kim SH Acta Crystallogr D Biol Crystallogr. 2004 May;60(Pt 5):903-11. Epub 2004, Apr 21. PMID:15103136[2]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
References
- ↑ Lesniak J, Barton WA, Nikolov DB. Structural and functional features of the Escherichia coli hydroperoxide resistance protein OsmC. Protein Sci. 2003 Dec;12(12):2838-43. PMID:14627744
- ↑ Shin DH, Choi IG, Busso D, Jancarik J, Yokota H, Kim R, Kim SH. Structure of OsmC from Escherichia coli: a salt-shock-induced protein. Acta Crystallogr D Biol Crystallogr. 2004 May;60(Pt 5):903-11. Epub 2004, Apr 21. PMID:15103136 doi:10.1107/S0907444904005013