User:Peggy Skerratt/Sandbox 1
From Proteopedia
[[Image:]]==RNA Polymerase==
This is a default text for your page Peggy Skerratt/Sandbox 1. Click above on edit this page to modify. Be careful with the < and > signs. You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
OverviewRNA polymerase is an enzyme that binds to double stranded DNA and synthesizes RNA. Since the enzyme moves along the DNA strand to synthesise it, it is considered a molecular machine. RNA polymerase is similar to the DNA polymerase because it reads the DNA strand from the 3’ end to the 5’ end. It synthesizes the RNA from the 5’ end to the 3’ end. It only reads the template strand to synthesize the complementary strand. Unlike the DNA polymerase it does not need a primer to begin, it can initiate synthesis itself. 1 This enzyme is found in both prokaryotes and eukaryotes. There are several different types of RNA polymerases. In Prokaryotes there is only one RNA polymerase, which synthesises all of the RNA. In eukaryotes there are three polymerases (RNA polymerase I,II, and III) that synthesize different types of RNA. Prokaryote and eukaryote polymerases have similar structures. Within the eukaryotic polymerases, the enzymes are structurally similar but have different subunits attached to the polymerase. The whole process of making RNA from DNA is called transcription. Transcription has four stages: assembly, initiation, elongation and termination, in which RNA polymerase is involved. The polymerase that is found to be a key polymerase is polymerase II. The exact mechanism for the different polymerases is still being studied. Since bacteria only have one polymerase that produce RNA, it is often studied to try to understand how this process works.The process of transcription is also regulated. It is controlled through specificity factors, repressors, and activators. If the transcription factors are not regulated, oncogenic activity can occur. Overall, RNA polymerase is still being studied and research is still being conducted to uncover all of the ins and outs of RNA polymerase’s role in transcription.
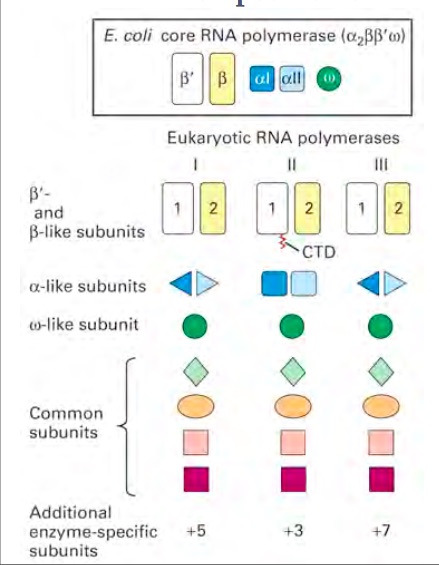
Structure and FunctionRNA polymerase is an enzyme that binds to double stranded DNA and synthesizes RNA. It is similar to the DNA polymerase because it reads the DNA strand from the 3’ end to the 5’ end. It synthesizes the RNA from the 5’ end to the 3’ end. It only reads the template strand to synthesize the complementary strand. Unlike the DNA polymerase it does not need a primer to begin, it can initiate synthesis itself. 1 There are different types of RNA polymerases that have different functions. The different polymerases all belong under the same family of enzymes. In eukaryotes and prokaryotes, the structures and mechanisms are similar to one another. However, they differ in the number of RNA polymerases that are used.1 In prokaryotes, there is only one type of polymerase that carries out all the functions that RNA polymerases have. The core RNA polymerase has different sigma factors linked to it that specify its binding to different promoter elements. The same process is more complicated in eukaryotes because they have different polymerases that transcribe different types of RNA.2 Eukaryotes utilize polymerases I, II, and III to form different types of RNA products. The RNA polymerase I produces ribosomal RNAs (rRNAs) in the nucleolus. Polymerase II produces messenger RNA (mRNA) in the nucleoplasm. Polymerase III produces tRNA and other small RNAs in the nucleus and cytosol. There are also other types of polymerases like Polymerases IV and V that are only found in plants.3 Even though different polymerases have different jobs their core structure is practically the same. The enzyme is composed of four different subunits. The four subunits are beta (β), beta prime (β′), alpha (α), and sigma (σ). These subunits have a molecular weight of 150,000, 160,000, 40,000, and 70,000 respectively. The sigma subunit is unique because it can dissociate from the complex. When the sigma subunit is dissociated, the enzyme is referred to as the core enzyme. The core enzyme is composed of two alpha subunits, one beta, and one beta prime subunit. However, when the sigma subunit is bound to the core enzyme, the enzyme is referred to as the RNA polymerase holoenzyme. It is this complete RNA polymerase holoenzyme that is vital to the first stage of transcription, the initiation step.4 In addition to this core structure there are enzyme specific subunits attached to each of the polymerases. Polymerase I contains five additional subunits, polymerase II contains three, and polymerase III contains seven addition subunits.5 There are also structural similarities in the eukaryotic polymerases. RNA Polymerase I and III are similar in the fact that they share two different alpha like subunits. This differs in Polymerase II because it has two copies of a different alpha like subunit. Polymerase II is unique because it contains a C- terminal domain (CTD). The phosphorylation of this CTD is vital for transcription and RNA processing.5 Research has come a long way in helping the world understand RNA polymerases; however, more research is currently being done to try to completely understand the ins and outs of RNA polymerases.
MechanismRegulationThough regulation is present throughout transcription it is especially important in RNA polymerase binding and transcription. Firstly, RNA polymerase only binds at the promoters of DNA. This binding at promoters can be controlled by specificity factors, repressors, and activators. Specificity factors can alter recognition and binding of the promoter. In Eukaryotes, the TATA binding protein is a specificity factor. Repressors, also known as negative regulators, bind to DNA at sites called operators. It is often near the promoter in bacterial cells. This causes a conformational change. Shock response during heat stress in E.coli is achieved by replacing the normal σ70 subunit is replaced by the σ32 specificity factor. Shock response can result in different proteins such as chaperones. Specificity factors can either increase or decrease transcription. Negative regulation is more common in bacteria, but eukaryotes such as yeast can use this mechanism. Often when eukaryotes use repressors they are often far from the promoter. Positive regulation is achieved through activators. They can bind to the DNA and enhance transcription when bound. Sometimes a signal molecule can cause an activator and the DNA to interact, causing an increase or decrease in transcription. cAMP receptor protein is an activator in E.coli. Activators are more common in eukaryotes. Architectural regulation is achieved by looping the DNA between the promoter and binding site. Connections with CancerRNA polymerase I transcription is promoted by tumor suppressors, oncogenes, and protein kinases. As a result disproportion between these promoters and RNA polymerase I is linked to cancer. RNA polymerase III products oversee traffic and synthesis of proteins. Transcription factors such as TFIIIB can interact with tumor suppressor genes. If Transcription factors are not regulated properly then oncogenic activity can occur.
| ||||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644