Sandbox RDE-1
From Proteopedia
Introduction
PMID: 23746446</ref>' scene='Insert optional scene name here' /> The rde-1 gene is a member of the Argonaute gene family. Proteins from "Argonaute" family form an evolutionarily conserved family whose members silence gene expression in pathways such as RNA interference (RNAi). Argonaute family proteins can be divided into two types, AGO and Piwi proteins, depending on the small RNA they bonded to. Both types of Argonaute proteins bind 21–35 nucleotide-long small RNA guides whose sequence identifies the genes to be silenced.[1] RDE-1 (RNAi-DEfective 1), a primary Argonaute protein, is required for RNA-mediated interference in Caenorhabditis elegans; thus, it is also known as RNAi promoting factor. Its gene locus was first characterized in C. elegans mutants resistant to RNAi, and was found to be a member of the Piwi gene family that includes plant, Drosophila, and vertebrate homologs.[2]
StructureAll "Argonaute"/Piwi proteins possess three primary domains forming a crescent-shaped base: the PAZ, MID, and PIWI domains. The amino-terminal PAZ domain uses its oligonucleotide-binding (OB) fold to secure the 3′ end of the small RNA guide strand to Argonaute protein. A conserved hydrophobic cavity within the PAZ domain recognizes the characteristic two-nucleotide, 3′ overhanging end of the guide-passenger siRNA generated by Dicer. The MID domain anchors the 5′ monophosphate of a siRNA to the Argonaute protein, securing the guide through multiple cycles of target cleavage. In vitro, studies suggest that 5′ phosphate binding helps align the small RNA on the surface of "Argonaute" protein, ensuring that the correct bond of the target is positioned in the endonuclease active site.[1] In other words, the PAZ and MID domains orient and anchor the double-stranded siRNA by binding to the 3’ and 5’ termini, respectively, leaving the internal nucleotides accessible for base pairing.[1][3] The carboxy-terminal PIWI domain resembles nuclease RNase H in which it folds into an RNase H-like structure. In RDE-1, this domain contains three conserved amino acids, aspartate-aspartate-histidine, that form a catalytic triad "DDH".[3] The crystal structure of RDE-1 has not been fully elucidated, but can be assumed to closely resemble its human homologs. The full length of RDE-1 protein is 1020 amino acids (aa)[2] in which about 110 of those aa makes up the PAZ domain and 300 aa makes up the PIWI domain.[4] Function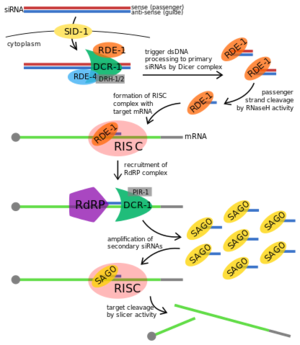 Proposed RNAi pathway for exogenous trigger dsRNA in C. elegans.[5] ImportanceMutation of any residues in the RNase H catalytic triad abolishes Slicer activity in human Argonaute protein Ago2, suggesting that the RNase H domain is directly responsible for target mRNA degradation.[11] However, RDE-1 has not been implicated in mRNA-cleavage activity. Instead, RDE-1 with mutations in the conserved DDH motif exhibit reduced passenger (sense) strand turnover, suggesting that RNase H activity serves to cleave the passenger strand, leaving the guide (antisense) strand accessible for base-pairing to target mRNA.[9] Further, target silencing can be fully restored in DDH motif mutants by loading single-stranded siRNA, suggesting that a downstream component in the RNAi pathway is responsible for Slicer activity.[9] Thus, RDE-1’s RNase H domain facilitates siRNA maturation but is not directly involved in cleaving target mRNA transcripts. RDE-1 is important for cleavage of the passenger strand and shuttling the siRNA to the appropriate RISC in C. elegans.[9][8] Without RDE-1, RISC cannot obtain the guide RNA for identifying the target transcript and thus transcript degradation cannot process properly or not at all. This protein is therefore also important for silencing a disease-causing gene by degradation of mRNA.[8] This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
References
| ||||||||||||
