Sandbox Reserved 1475
From Proteopedia
</StructureSection>
| This Sandbox is Reserved from November 5 2018 through January 1, 2019 for use in the course "CHEM 4923: Senior Project taught by Christina R. Bourne at the University of Oklahoma, Norman, USA. This reservation includes Sandbox Reserved 1471 through Sandbox Reserved 1478. |
To get started:
More help: Help:Editing |
This page is reserved for Marisha
Contents |
Retinal Dehydrogenase Type Two Structure
|
This molecular structure is Retinal Dehydrogenase Type Two (RalDH2) which was extracted from Rattus norvegicus.[1] The sample was a crystallization of a clone of RalDH2 that was given as a gift from J. L. Napoli. It has been shown that RalDH2, the cellular binding protein type I, and the retinol binding protein receptor all co-localize in tissues that require Vitamin A (retinol) for normal development in rat embryos post-gastrulation, as discussed by "personal communication" to the authors of the "The Structure of Retinal Dehydrogenase Type II at 2.7 Angstrom Resolution: Implications for Retinal Specificity".[1] This enzyme is part of the super family Aldehyde Dehydrogenase. The function of this enzyme is to catalyze the oxidation of retinal to retinoic acid. Retinoic acid produces a putative morphogen that initiates pattern formation in the early embryo.[1] This is the last step in the formation of the hormone from Vitamin A (retinol).[1] Vitamin A that has been metabolized can produce retinoid derivatives which function in either vision or growth and development. [2] RalDH2 is expressed in Escherichia coli strain BL21(DE3) [3]
Function
The main function of this enzyme is to Retinoic acid. RalDH2 requires (NAD+) as a cofactor.[1] In the oxidoreductase reaction, NAD+ acts as an electron acceptor. The reaction of this enzyme is [(retinal) + (NAD+) + (H2O) ↔ (retinoic acid) + (NADH) + (H+) ]. Once the NAD+ is bound, hydrogen bonds form with non-polar residues and one basic Lysine residue. Chloride ions participate in hydrophobic interactions with Arginine residues.[1] Theres interactions cause a structural change to occur in the RalDH2 enzyme which causes it to form a more favorable folded confirmation. In the enzyme a large binding cavity is formed. tructural changes occur to stabilize the tertiary structure of RalDH2
Structural highlights
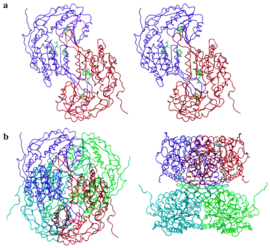
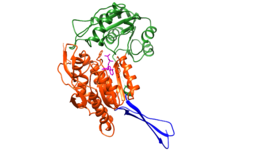
The structure was based on the mitochondrial aldehyde dehydrogenase type two. RalDH2 in a monomer made up of 3 domains: a nucleotide-binding domain (1-136, 161-270), a catalytic domain (271-484), and a tetramerization domain (137-160, 485-484) as shown in Figure 1.[1] The tetramer can be envisioned as an "X", with nucleotide-binding sites at the tips of the "X", and the tetramerization domains as the equatorial portion of the "X" as seen in Figure 2.[1] In Figure 3 it is possible to see the active site, which is where the substrate interacts with Cys-302.
</StructureSection>
Substrate NAD
The crystal structure was cocrystallized with , and was determined at a 2.7 Angstrom resolution. [1]
Disease
Relevance
</StructureSection>
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Lamb AL, Newcomer ME. The structure of retinal dehydrogenase type II at 2.7 A resolution: implications for retinal specificity. Biochemistry. 1999 May 11;38(19):6003-11. PMID:10320326 doi:10.1021/bi9900471
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedFamilies_of_Retinoic_Dehydrogenases - ↑ Lamb AL, Wang X, Napoli JL, Newcomer ME. Purification, crystallization and preliminary X-ray diffraction studies of retinal dehydrogenase type II. Acta Crystallogr D Biol Crystallogr. 1998 Jul 1;54(Pt 4):639-42. PMID:9761861