Sandbox Reserved 1475
From Proteopedia
</StructureSection>
| This Sandbox is Reserved from November 5 2018 through January 1, 2019 for use in the course "CHEM 4923: Senior Project taught by Christina R. Bourne at the University of Oklahoma, Norman, USA. This reservation includes Sandbox Reserved 1471 through Sandbox Reserved 1478. |
To get started:
More help: Help:Editing |
This page is reserved for Marisha
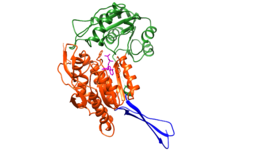
Retinal Dehydrogenase Type Two Structure
|
This molecular structure is Retinal Dehydrogenase Type Two (RalDH2) which was extracted from Rattus norvegicus.[1] This enzyme is part of the super family Aldehyde Dehydrogenase. The sample was a crystallization of a clone of RalDH2 that was given as a gift from J. L. Napoli. It has been shown that RalDH2, the cellular binding protein type I, and the retinol binding protein receptor all co-localize in tissues that require Vitamin A (retinol) for normal development in rat embryos post-gastrulation, as discussed by "personal communication" to the authors of the "The Structure of Retinal Dehydrogenase Type II at 2.7 Angstrom Resolution: Implications for Retinal Specificity".[1] RalDH2 is the enzyme that produces retinoic acid in mouse embryos during gastrulation, and is found to be surrounding the primitive node, which is where retinal is converted into retinoic acid.[2] The function of this enzyme is to catalyze the oxidation of retinal to retinoic acid. Retinoic acid produces a putative morphogen that initiates pattern formation in the early embryo.[1] This is the last step in the formation of the hormone from Vitamin A (retinol).[1] Vitamin A that has been metabolized can produce retinoid derivatives which function in either vision or growth and development.[3] RalDH2 is expressed in Escherichia coli strain BL21(DE3) [4]
Function
The reaction of this enzyme is [(retinal) + (NAD+) + (H2O) ↔ (retinoic acid) + (NADH) + (H+)]. The main function of this enzyme is to produce Retinoic acid. RalDH2 requires (NAD+) as a cofactor.[1] In the oxidoreductase reaction, NAD+ acts as an electron acceptor. Once the NAD+ is bound, hydrogen bonds form with non-polar residues and one basic Lysine residue. Chloride ions participate in hydrophobic interactions with Arginine residues.[1] Theres interactions cause a structural change to occur in the RalDH2 enzyme which causes it to form a more favorable folded confirmation. In the enzyme a large binding cavity is formed. tructural changes occur to stabilize the tertiary structure of RalDH2