Dihydropyrimidine Dehydrogenase (DPD) is an important enzyme involved in the degradation of pyrimidines in the body. It is a 220 kDa protein that binds co-factors; FAD, FMN, [4Fe-4S] clusters, and substrates; NADPH and pyrimidines.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Function
Dihydropyrimidine Dehydrogenase (DPD) is the first enzyme in pyrimidine degradation pathway. It catalyzes the reduction of 5,6-double bond to obtain dihydropyrimidine.
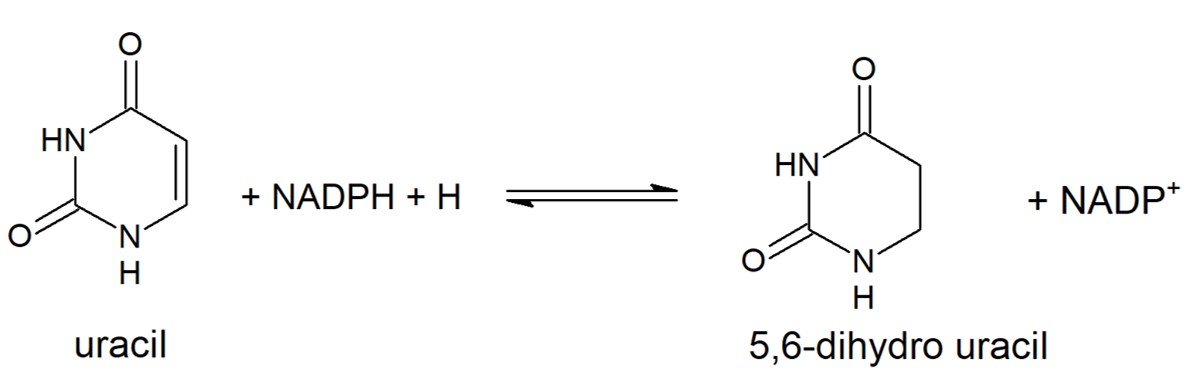
Reduction of Uracil to form 5,6-dihydro uracil. This reaction is catalyzed by eukaryotic DPD. Other pyrimidines can take the place of uracil in this reaction and will be metabolized in the same way.
Disease
5-fluorouracil (5-FU) is a drug used to treat a variety of cancers as it has wide anti-tumor activity & works well alongside other chemotherapy drugs. In the human liver 80-85% of 5-FU is catabolized into inactive, and potentially toxic, metabolites by DPD.
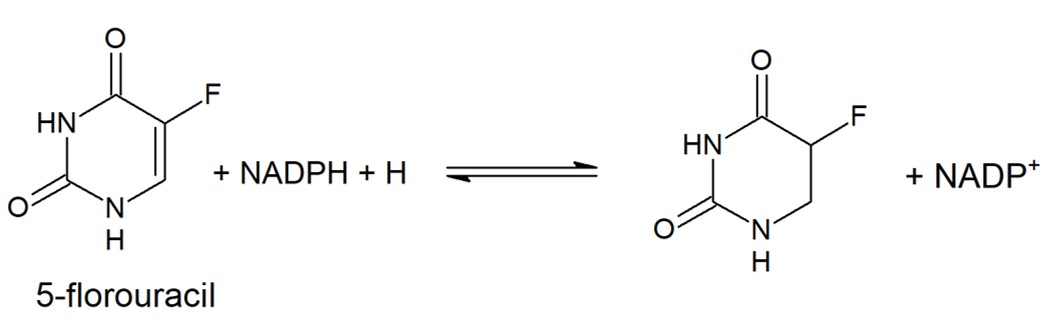
Only 1-3% of the original dose proceeds through anabolic pathways to create active cytotoxic complexes. The active complexes inhibit DNA synthesis and the processing and function of RNA processing thus producing a deleterious effect on both healthy and cancerous cells.
DPD decreases effectivity of drug thus requires a very high dosages, leading to major side effects. Luckily, inhibitors are in development and some are in clinical trials.
Structural highlights
DPD is a homodimer, with each monomer consisting of five domains.
Domain 1
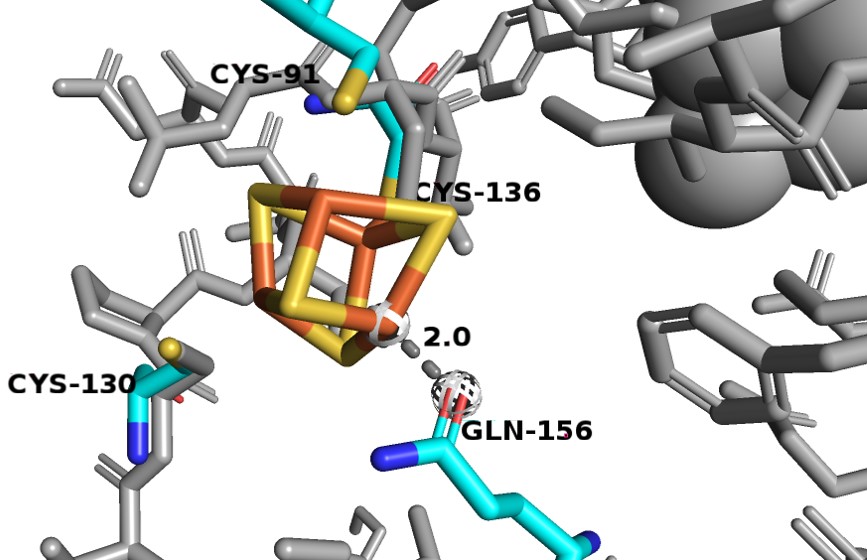
Consists of residues 27-173. It binds 2 Fe-S clusters and is made of exclusively alpha-helices. One of the [4Fe-4S] clusters is odd as it has a different coordination that is not observed in other Fe-S proteins. Three Fe atoms interact with cysteines 91, 130 and 136, but the fourth Fe atom interacts with the Oε1 glutamine 156. Since the oxygen has a smaller atomic radius than sulfur, so the Fe-O distance is much shorter (~2.0 Å) than the average Fe-S interactions seen in DPD (2.3 Å).
Domain 2 and 3
Domain 2 consists of residues 173-286 and 442-524. It binds FAD and NADPH. It contains a central parallel beta-sheet surrounded by alpha helices, forming Rossman-type nucleotide binding motifs that bind FAD and NADPH.
Domain 3 consists of residues 287-441 and also binds FAD and NADPH. Just like domain 2, contains a central parallel beta-sheet surrounded by alpha helices, forming Rossman-type nucleotide binding motifs that bind FAD and NADPH. But, D3 also contains an additional 3 stranded antiparallel beta-sheet. With the aforementioned exception, D2 and D3 are so similar that they are thought to have originated from gene duplication.
In the following figure, domain 2 is shown in blue, domain 3 is in green, FAD is colored in orange and NADPH is in pink.



