This is a default text for your page '. Click above on edit this page' to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Function(s) and Biological Relevance
Lignostilbene-α,β-dioxygenase A (LsdA) from the bacterium Sphingomonas paucimobilis TMY1009 is a nonheme iron oxygenase that catalyzes the cleavage of lignostilbene, a compound arising in lignin transformation, to two vanillin molecules. LsdA has greatest substrate specificity for lignostilbene. The substrate's 4-hudryoxy moiety is required for catalysis. Phenylazophenol inhibits the cleavage of lignostilbene by LsdA. The breaking down of lignin is essential to the sustainable biorefining of lignocellulose. It is of great relevance to transforming lignocellulose to biofuels.
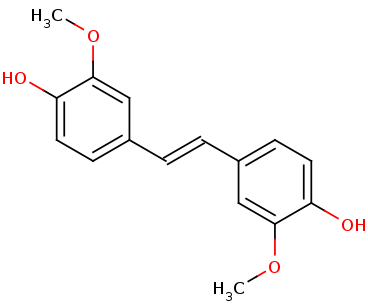
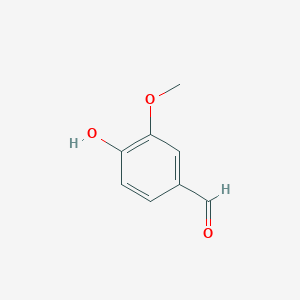
Broader Implications
Lignin represents 30% of the lignocellulose biomass. It consists of different aromatic building blocks, phenylpropanoids, which are extremely useful. Normally aromatic compounds are extracted from petroleum and are used to manufacture drugs, paint, plastics, etc. Therefore the potential of lignin is very high. Lignin is the most abundant polymer in nature other than cellulose and chitin, and it is the only one that contains such a large number of aromatic compounds.
Structural highlights and structure-function relationships
The flat surface of the B-sheet is pushing the amino acids up, making it possible for the catalytic triad Phe59, Tyr101, and Lys134 to create interactions with the ligand.
[[Image:]]
Energy Transformation
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.


