This is a default text for your page Elizabeth M. Ratz/Sandbox 1. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Introduction
Calcium is a very important signaling molecule in the body with many physiological functions including muscle contraction, neuron excitability, cell migration and growth. The mitochondria are important regulators of calcium in the body and the calcium uniporter (MCU) maintains calcium homeostasis within the mitochondria. Calcium moves in one direction from the intermembrane space through the inner mitochondrial membrane into the matrix. The matrix is more negative driven by the respiratory chain which draws calcium in and allows calcium to move down its gradient.
The MCU is a complex. Its MICU1 and MICU2 bind together and associate with EMRE which regulates MCU. The MICU1 and MICU2 act as gatekeepers. EMRE connects the MICU1 and MICU2 sensors to MCU therefore regulating calcium uptake for the protein
The selectivity pore is an integral part of the protein. This pore contains a group of glutamate with oxygen facing inward forming a carboxylate ring through which calcium enters. This negative carboxylate ring does a good job of pulling the positive calcium into the selectivity pore at the top of the protein.
Cryogenic electron microscopy (Cryo-EM) was instrumental in outlining the complete structure of this protein.
Structural highlights and mechanism
The MCU is a dimer of dimers, described as tetrameric truncated pyramid. The uniporter has only a single strong binding site located in the selectivity pore with specificity for
Calcium, near the surface of the inner mitochondrial membrane. Activity of the uniporter is dependent on membrane potential and calcium concentration. Calcium from the cytoplasm enters the mitochondrial innermembrane space through the mitochondrial membrane and is passed to the mitochondrial matrix via the MCU.
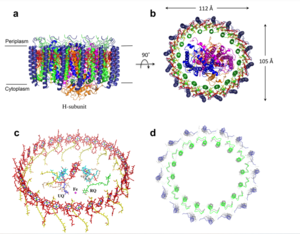
Figure 1: structure of mitochondrial calcium uniporter
Transmembrane Domain
The is on the inner mitochondrial membrane open to the inner membrane space. The small pore, highly specific for calcium binding is located in (TM2) while (TM1) surrounds the pore. The transmembrane domain exhibits four fold rotational symmetry. It is important that the selectivity pore is small, allowing only a dehydrated calcium molecule to interact with the 5 ampier wide glutamate ring. The negative charge of the glutamates carboxyl group attracts the positively charged Calcium ion. Approximately one helical turn below the glutamate ring of the selectivity filter, there is a tyrosine ring coming a 12 ampier wide pore allowing high conductivity. The wider opening allows calcium to rehydrate once they pass the selectivity pore. The domain swapping of TM1 of one subunit with the TM2 of the neighboring subunits allows for a tight packing in the transmembrane connectivity providing flexibility to the uniporter.
Soluble Domain
The is the first subsection of the soluble domain, which resides in the inner mitochondrial membrane. The coiled coil functions as the joints of the uniporter, providing flexibility to promote transport of Calcium ions down their concentration gradient. The junction between the transmembrane domain and the coiled coil's flexibility can be attributed to the disordered packing between subunits; subunits A and C adopt different conformations than the B and D subunits, although they superimpose well.
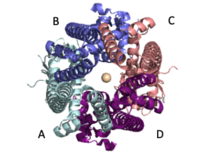
Figure 2: Symmetry and organization of subunits from looking down into the uniporter from the inner mitochondrial membrane
When calcium binds to the selectivity pore, the coiled coil swings approximately 8 degrees around its end near the . This movement propagates to the top of the transmembrane domain, where the pore is located, about 85 amperes away. The largest displacement triggered by the movement of the coiled coil is in the transmembrane domain, where the coil bends 20 degrees, moving the transmembrane domain further apart. The N-Terminal domain (NTD) is involved in calcium condition. Reorganization in the NTD due to shifts in the coiled coil switch subunits to alter membrane packing causing the interactions between the tyrosines and transmembrane helices. This propagation facilitates a rotamer switch between one pair of tyrosine controlling calcium flow through the pore. The soluble domain is wider than the transmembrane domain, allowing calcium ions to rehydrate and increasing the conductivity of ions through the uniporter into the mitochondrial matrix.
Selectivity Filter
The contains Glu358, Trp354, and Pro359 to allow calcium to pass through the uniporter. The carboxylate oxygen of the side chains draw in the positive calcium ion. The of the carboxyl ring is about 4Å, allowing only a dehydrated Ca ion to bind. Trp38, which is directly next to the Glu residues, stabilizes the carbonyl side chains through and anion pi interactions. These Trp residues also form stacking interactions with Pro359, which orientate the Glu carboxyl side chains towards the middle of the pore to interact with Ca ions.
Calcium Uniporter Structure
[3]
Function
Inhibitors

Figure 3: Competitive inhibitors Ruthenium Red (A) and Gadolinum (B)
Disease Links
Types 1 and 2 Diabetes
Pancreatic beta cells circulate insulin through the body. Glucose initiates signals allowing these cells to break down sugar and release insulin, which is all stimulated by mitochondrial energy metabolism. Calcium homeostasis plays a fundamental role in ATP production supplying energy to this process. Defects in homeostasis of calcium like chronic calcium depletion, caused by leaky Ryanodine receptors, causes types 1 and 2 diabetes through failure of this mechanism. Treatment involves targeting the MCU and MICU1 to open the calcium channel and allow more uptake of calcium ions into the mitochondria.
Heart Failure
Calcium overload in the mitochondria of cardiac cells lead to apoptotic cardiac cell death. This large amount of cell death
Relevance
Structural highlights
Student Contributors
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.



