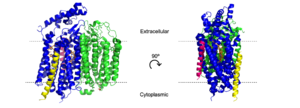Introduction
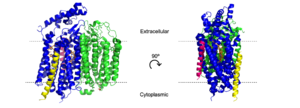
'
Figure 2. Cartoon model of cytochrome bd-oxidase in
E. coli. Dashed lines represent borders of cytoplasmic and extracellular regions.
bd Oxidase is a type of quinol-dependent terminal oxidase found exclusively in prokaryotes.[1] With a very high oxygen affinity, bd oxidases play a vital role in the oxidative phosphorylation pathway in both gram-positive and gram-negative bacteria. bd oxidases responsibility in the oxidative phosphorylation pathway allows the protein to also assist as a key survival factor in the bacterial stress response against antibacterial drugs. [2] The allows bacteria to be resistant to hypoxia, cyanide, nitric oxide, and H2O2[3]
Given this knowledge, bd oxidases have become an area of scientific research worth pursuing as they could serve as an ideal target for antimicrobial drug development.
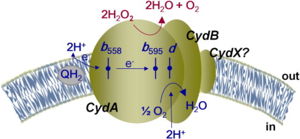
Figure 1: Overall schematic representation of cytochrome bd.
[4]; General display of the reduction of molecular oxygen into water using the quinol as a reducing substrate. The three hemes are located near the periplasmic space, meaning that the membrane potential is generated mainly from proton transfer from the cytoplasm towards the active site on the opposite site of the membrane. Heme
b558 is involved in quinol oxidation and Heme
d serves as the site where O2 binds and becomes reduced to H2O.
The overall mechanism of bd oxidases involves an exergonic reduction reaction of molecular oxygen into water (Figure 1). During this reaction, a proton gradient is generated in order to assist in the conservation of energy. Unlike other terminal oxidases, bd oxidases do not use a proton pump. Instead, bd oxidases use a form of vectorial chemistry that releases protons from the quinol oxidation into the positive, periplasmic side of the membrane. Protons that are required for the water formation are then consumed from the negative, cytoplasmic side of the membrane, thus creating the previously mentioned proton gradient.
This page will be specifically focusing on the structure and overall function of the 6RX4 bd oxidase. 6RX4 is a part of the long(L) quinol-binding domain subfamily that terminal oxidases are classified into. The L-subfamily of bd oxidases are responsible for the survival of acute infectious diseases such as E.Coli and salmonella. The 6RX4's three groups, its periplasmically exposed , and will be of primary focus when identifying the relationship between structure and function.
Structure
Subunits
E. coli bd oxidase is made up of four individual subunits.[5] The two major subunits, CydA and CydB, are each composed of one peripheral helix and two bundles of four transmembrane helices. The plays the most important role in the oxygen reduction reaction as it contains the Q-loop as well as all three heme groups. The harbors the molecule which provides structural support to the subunit that mimics the three hemes found in CydA.[1][6] The remaining two subunits, CydS and CydX, are both single helix structures that assist in the oxygen reduction reaction. Unique to E. coli, the binds to CydA to block oxygen from directly binding to heme b595. The promotes the assembly and stability of the oxidase complex. CydX is composed of 37 mostly hydrophilic amino acid residues, including that is exposed to the cytoplasm and prevents the helix from fully entering the membrane. [5]
Q-Loop
Another significant structural feature of bd oxidase is the which is located between TM helices 6 and 7 of the CydA subunit.[5] The periplasmic Q-loop in E. coli stretches over a length of 136 amino acid residues, making it much longer than the Q-loop in Geobacillus thermodentrificans.[1] The Q-loop is likely involved in quinone binding and oxidation. The N-terminal end of this Q-loop is very flexible and likely functions as the hinge that allows for quinone binding while the C-terminal end is much more rigid which provides stabilization for the enzyme.[5]
Molecular Function
H and O channels

Figure 3. H and o-channels of cytochrome bd-oxidase in
E. coli. Channels are outlined in gray, water is shown as spheres, and various amino acids are labeled above.
The hydrogen and oxygen channels (Fig. 3) are essential for H+ and O2 molecules to reach the active site of cytochrome bd oxidase. A proton motive force generated by the oxidase[1] allows protons from the cytoplasm to flow through a hydrophilic full of water (pink dots), entering at and moving past [5] where they can be transferred to the active site with the help of the conserved residues [1]. A smaller also exists that transitions from hydrophobic to hydrophilic as it gets closer to the active site. This channel allows oxygen to reach the active site, starting near in CydB and passing by [1], which assists with the binding of oxygen to the active site. The o-channel channel is approximately 1.5 Å in diameter[5], which may help with selectivity.
Interestingly, the o-channel does not exist in the cytochromebd oxidase of Geobacillus Thermodenitrificans; instead, oxygen binds directly to the active site[6]. The subunit found in E. coli blocks this alternate oxygen entry site, which allows oxygen to travel through the o-channel[1][5]. The presence of an o-channel affects oxidase activity, as the E. coli oxidase acts as a "true" oxidase, while the G. th bd oxidase contributes more to detoxification[5].
Hemes
There are three molecules present in the CydA subunit that form a triangle to maximize subunit stability[5][1][6], which is an evolutionary conserved feature across bd oxidases[1]. Similar to the hemes, the (UQ-8) molecule found in CydB mimics the triangular formation to stabilize the subunit[1]. Heme b558 acts as the primary electron acceptor by catalyzing the oxidation of quinol[5]. Conserved help to stabilize heme b558[5]. Heme b558 transfers the electrons to heme b595, which transfers them to the active site heme d[1]. A conserved assists heme b595 in transferring electrons to heme d[6]. A conserved is essential for charge stabilization of heme b595[5], while stabilizes heme d[6]. As heme d collects the electrons from heme b595, in the o-channel facilities the binding of oxygen to heme d, and in the h-channel facilitate proton transfer to heme d[1]. With electrons, oxygen, and protons available, heme d can successfully reduce dioxygen to water.
Relevance
The cytochrome bd oxidase is essential for bacteria to thrive in the human body. Terminal oxidases in bacteria are needed for formate oxidation activity, which provides a sustainability advantage for bacterial growth. If E. coli are missing or possess ineffective CydA and B subunits, their advantage is eliminated[7]. Specifically with colitis, E. coli mutants that were missing CydAB colonized quite poorly, while the wild type colonized at high levels[7]. The cytochrome bd oxidase is the main component in nitric oxide (NO) tolerance in bacteria, which is released by neutrophils and macrophages when the host is infected[8]. E. coli growth seen in urinary tract infections is mainly due to the NO resistant bd oxidase, but without the CydA A and B subunits, bacteria cannot colonize in high NO conditions[8]. Cytochrome bd oxidases are essential in other bacteria, specifically in M. tuberculosis. Other known oxidases can be inhibited to prevent the spreading of M. tb, however the cytochrome bd oxidase not only allows M. tb to survive, but to colonize. Without the CydAB subunits, M. tb growth dramatically decreases when exposed to imidazo[1,2-α]pyridine, a known inhibitor of ATP synthase[9].
Due to that fact that it is only found in prokaryotes and considering its relevance in notable bacterial infections, inhibitors that target cytochrome bd oxidase are quite practical. Compounds that target heme b558[3], create unusable forms of oxygen[10], and target the o-channel [11] have shown tremendous potential in halting bacterial growth.