User:Fujr Ibrahim/Sandbox 1
From Proteopedia
Contents |
Miraculin, a taste-deceiving protein
This is a default text for your page Fujr Ibrahim/Sandbox 1. Click above on edit this page to modify. Be careful with the < and > signs. You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Introduction to Miraculin
Miracle? I think you mean Miraculin. Miraculin is a protein that is best known for its ability to deceive human taste buds into thinking sour or acidic food is sweet. This homodimeric glycoprotein was first identified in the West African native fruit, Synsepalum dulcificum (also known as “Miracle Fruit”), and exists in the pulp of the miracle fruit.
Miraculin’s deceptive properties have been exploited by several companies as sugar substitutes. However, the American Food and Drug Administration banned the use of miraculin after labeling it as an additive [3] . This prevented its commercial use in the food industry.
Miraculin’s Structure
Miraculin is composed of 191 amino acid residues linked primarily by peptide bonds, having a molecular weight of about 28 kDa. Sarroch Theerasil et al [4] use HPLC profiles and SDS-PAGE analyses to prove this.
Miraculin is a homodimer made by two chains that have two N-glycosylated Asn residues and are cross-linked through a disulfide bridge. Miraculin can also exist in a tetramer form.
Predictivley-modeled structure of miraculin
Antonella Paladino et al’s article “Molecular modeling of miraculin: Structural analyses and functional hypotheses” [5] models the structure of miraculin by comparative modelling and molecular docking techniques as no structural data for miraculin is available. The article concludes that two histidine residues, located in exposed regions, are the main responsible of miraculin activity. The assays run in the journal also conclude that the miraculin dimer assumes a widely open conformation in an acidic environment. Although not directly apparent in the model, four hydrogen bonds are present between the two dimers. In linear form, only one hydrogen bond is possible between the dimers. The different conformations can be used to compare the closed and open conformations of miraculin in figure 2 of the journal. The prevalence of cysteine and histidine residues are highlighted in the figure as well.
Miraculin’s interactions with human tongue receptors
Taste processing is a complex process and is initially achieved by the activation of taste receptor cells clustered on the tongue’s taste buds. Once activated by a wide variety of ligands, the taste receptor cells transmit signals to parts of the brain that are involved in taste perception [6] Like many interactions involving the binding of a ligand to a receptor, miraculin undergoes a conformational change when binding to the tongue receptors where its active site shifts to better bind to tongue receptors [7]. Miraculin binds to the tongue’s HT1R2-HT1R3 (human taste type 1 receptor 2 and 3) receptors in a pH-dependent manner. HT1R2-HT1R3 is a G-protein coupled receptor that is also capable of binding to natural sugars and artificial sweeteners. Recent studies suggested also that the association of the closed and open forms of monomers constituting the T1R2 T1R3 heterodimer can create a large charged cavity where sweet proteins fit exerting their function[8]. Interestingly, although miraculin is inactive at very basic conditions, it still capable of suppressing the response of HT1R2-HT1R3 to other sweet-tasting compounds at neutral pH. At acidic conditions, miraculin enhances HT1R2-HT1R3’s response to sweet-tasting compounds. [9] Two histidine residues, His30 and His60, participate in the process of taste-modification. One site maintains the attachment of the protein to the membranes while the other activates the sweet receptor membrane in acidic conditions. [10] Although the detailed mechanism of the taste-deceiving protein is unknown, numerous sources agree to the fact miraculin is activated in the presence of an acidic compound. At an acidic pH, miraculin assumes an open conformation that permits it to bind to the tongue’s HT1R2-HT1R3 receptors. Miraculin is denatured at high temperatures and at pHs below 3 or above 12. The denaturation at these conditions is most likely due to the loss of shape of the protein and the disruption of bonds critical to its functionality.
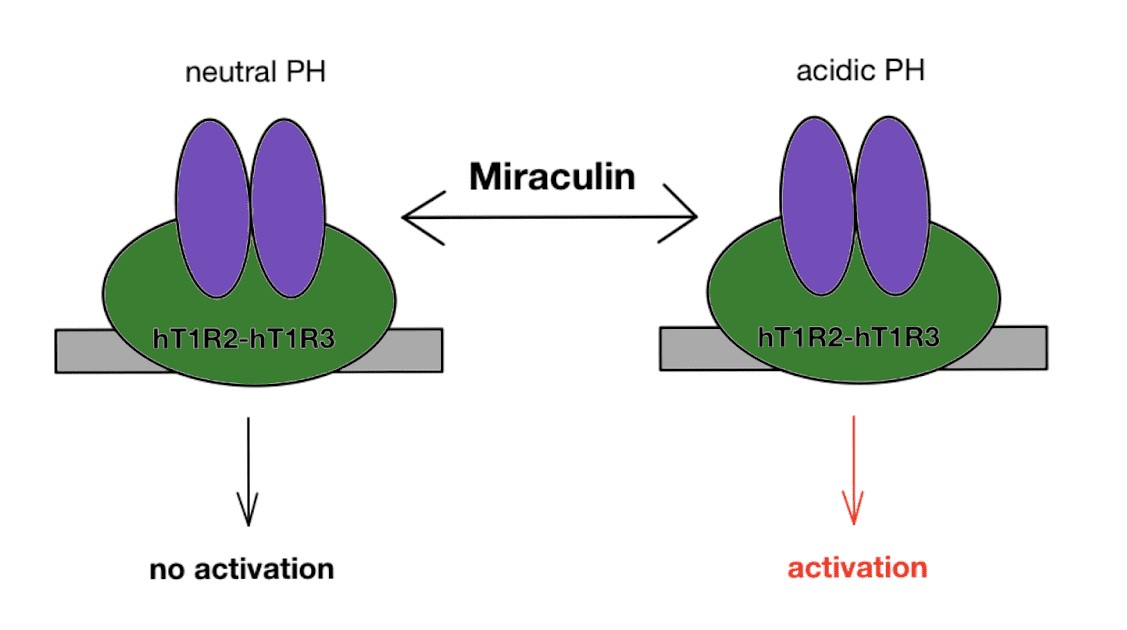 Fig 1. Miraculin activates taste receptor, HT1R2-HT1R3 at acidic pH
Fig 1. Miraculin activates taste receptor, HT1R2-HT1R3 at acidic pH
Miraculin-Like Proteins (MLPs)
Several miraculin-like proteins (MLPs) have been identified and are classified as “miraculin-like” based on amino-acid sequence alignment with that of miraculin. MLPs and miraculin are categorized into the Kunitz-type soybean trypsin inhibitor (STI) family. Features common to the Kunitz-type soybean trypsin inhibitor (STI) family include the presence of disulfide bridges --which are apparent in miraculin models-- and the inhibition of trypsin and chymotrypsin. Examples include MLPs extracted from Murraya koenigii and Vitis vinifera plants. Since atomic-level structural data of miraculin is not available to date, A miraculin-like protein (MLP) homologous to miraculin extracted from Murraya koenigii will be used for protein visualization purposes.
MLP Extracted from Murraya koenigii (Heterodimer)
This MLP consists of 190 amino acid residues with seven cysteines arranged in three disulfide bridges and has a mass of 21.4kDa. Crystal structure analysis in figures 2a and 2b shows that the protein is composed of antiparallel beta-strands, loops that connect the beta-strands, and 4 helix turns (fig2b). Figure 2a shows that are 6 disulfide bridges that hold the monomers together in a manner similar to that between monomers in miraculin.
| |||||||||||
