User:Dora Bonadio/Sandbox 1
From Proteopedia
SARS-CoV-2 main protease (Mpro)
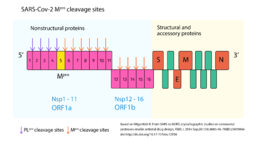
IntroductionThe (Mpro) protease (also known as 3CLpro), is a viral non-structural protein from the virus SARS-CoV-2 [1], responsible for a major outbreak of the disease called Coronavirus Disease 2019 (COVID-19), declared pandemic by WHO in 11 march 2020 [2]. It has an important role in virus replication, as it’s responsible for cleavage of the polyproteins of the virus, alongside with papain-like protease(s) [3]. FunctionThe (Mpro) protein function is mainly deduced from the function of SARS-CoV virus (Mpro), which has a 96% amino acid identity and a highly similar three-dimensional structure with SARS-CoV-2 (Mpro) [1]. As a protease, (Mpro) is an enzyme that causes proteolysis, which means that it breaks protein peptide bonds by hydrolysis [4]. Indeed, the (Mpro) processes the replicase polyprotein 1ab (pp1ab ~790 kDa) translated from the viral RNA ORF1ab [1][5]. In fact, (Mpro) cleaves 11 sites of pp1ab and the recognition sequence at most sites is between Leu-Gln and (Ser, Ala, Gly) [1][3]. Proteins resulting from this polyprotein cleavage are non-structural proteins (NSPs) and they seem to contribute with viral replication and transcription [5]. Thus, by processing an important number of non-structural proteins, this enzyme plays a critical role in SARS-CoV-2 replication. StructureThe (Mpro) is a protein of approximately 30 kDa [5][6] consisting of containing 306 amino acid residues each [1]. This protomers dimerize forming a homodimer [1]. Each protomer consists of : I (; residues 10-99), II (; residues 100-182), and III (; residues 198-303). Domains I and II comprise six-stranded antiparallel β-barrels and domain III comprises five α-helices [1][6]. The substrate-binding site is located between domains I and II with the containing the amino acid residues Cys145 and His41 [1]. Domain III, in turn, has been shown to be involved in the regulation of (Mpro) dimerization, what is necessary for the catalytic activity of this enzyme once it helps to shape the substrate-binding site [1][7]. Structural comparison with SARS-CoV MproAs mentioned above, SARS-CoV-2 (Mpro) has 96% sequence identity with SARS-CoV (Mpro) and as expected, also a highly similar three-dimensional structure [1]. Indeed, it has been shown that the substrate-binding pocket is a highly conserved region of (Mpro) among an important number of CoV (Mpro) [6]. However, an interesting difference found between SARS-CoV (Mpro) and SARS-CoV-2 (Mpro) is that in the first one there is a polar interaction between the domains III of each protomer, involving the residues Thr285, what is not found in the COVID-19 virus (Mpro) [1]. In fact, in SARS-CoV-2, the threonine is replaced by , leading to a higher proximity between the two domains III of the dimer [1]. An attractive drug targetAs have been shown, because of its importance for viral replication, inhibiting SARS-CoV-2 (Mpro) activity could lead to viral replication blockage [1][6]. Moreover, no human proteases has been reported to have a similar cleavage specificity and so, in this aspect, (Mpro) inhibitors toxic side-effects may be reduced [3]. Therefore, CoV (Mpro) has been an attractive drug target among coronaviruses [3] and so it is for COVID-19 [1][6]. Indeed, virtual drug screening, structure-assisted drug design, and high-throughput screening are been used to repurpose approved pharmaceutical drug and drug candidates targeting SARS-CoV-2 (Mpro) [6][8]. Furthermore, a study carrying the pharmacokinetic characterization of an optimized (Mpro) provides useful framework for development of this kind of inhibitors toward coronaviruses [1]. It was showed that the α-ketoamide inhibitor interacts with the catalytic site of the enzyme through two hydrogen bonding interactions, as can be seen in the complex formed between SARS-CoV-2 (Mpro) and an [1]. External Resources
References
| ||||||||||||