This is a default text for your page Andre Wu Le Chun/Sandbox 1. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Structural highlights
6svb is a homotrimer transmembrane glycoprotein(S) protruding from the viral surface¹. It is composed by two subunits that are fundamental for the virus entry in the cell: The S1 subunit, which contains the receptor binding domain (RBD), is responsible for binding to the host cell receptor, and the S2 subunit, responsible for fusion of the viral and cellular membranes. In order to allow the binding of the RBD in S1 with the host cell, the spike protein goes through conformational changes that start from a pre-fusion conformation to a stable post-fusion conformation, in which the receptor-binding domain assumes a up conformation.
This protein also shares amino acid sequence identity of 76% with SARS-CoV, however it has inserted amino acid that indicate a furin like cleavage site in the S1/S2 boundary, that must be primed in order to enable the viral pathogenicity. In the Receptor binding domain, units of beta-sheets, alpha-helices and loops can be observed.
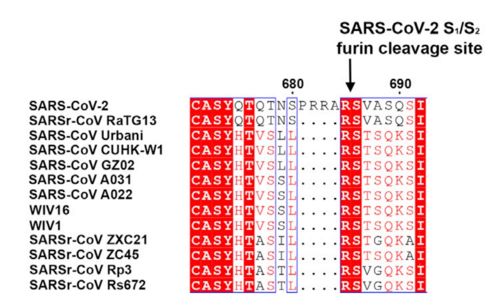
figure 1: Alignment of S in between multiple SARS-CoV (Walls et al., 2020)
Function
The spike glycoprotein binds the viral structure to the hosts cell's angiotensin-converting enzime 2 (1r42). Once bound, it starts the fusion of viral and cellular membrane, therefore enabling the pathogenicity and spread of the virus.
Disease
6vsb is probably the protein that enabled the Sars-Cov-2 to enter human cells, rusulting therefore on the large scale coronavirus epidemics in 2020.
As the name implies, The main symptoms of the COVID-19 were similar to the ones of common cold: fever, cough and tiredness, but with some stronger than the common cold: Chest pain, lost of smell and taste sense.
The main reason for the worst symptoms are the death of several types of cells in the lungs, causing severe loss of oxygenation, therefore resulting of gradual colapse of the respiratory system of the infected.
Relevance
Due to its role in the infection process, the spike glyprotein may be a potential target of studies that seek methods of preventing the COVID-19 disease. These include the development of vaccines based on the protein's structure of the protein and on it's recognition/biding mechanisms. For instance, avoiding the cleavage of the furin, located on the B domain of the protein, by the host's proteases could be a way of viral inhibition. Comprehending how the protein can be bound to antibodies could also help the scientific community to prepare for future SARS coronavirus outbreaks that may happen in the future.
Interaction with angiotensin-converting enzime 2
The interaction between the 2019-nCov and the host cell begins with the recognition of the ACE2, a dimeric protein. Then, the S1 subunit moves, modifying the protein's conformation in way that receptor-binding domain becomes available to connect itself to the peptidase domain of target cell. Due to the existence of four amino acids in between the S1 and S2, a furin cleavage site is featured in the protein. The cleavage of this site has potential relevance as it could lead to later priming at the S2' site by a serine protease, TMPRSS2. Once attached on the surface of ACE2, the cleavage of S2' happens and the fusion mechanism is activated. Then, the protein goes through conformational changes, the protein structure assumes a conformation which is suitable for binding with the ACE2 receptor. At that instance, the spike protein is found in a "up" conformation, hence the protein's name. It binds itself to the ACE2 with high affinity (15nM). In order to activate infection, the spike protein is cleaved by TMPRSS2, a host's serine protease, also fundamental for the viral infection. Furine cleavage site
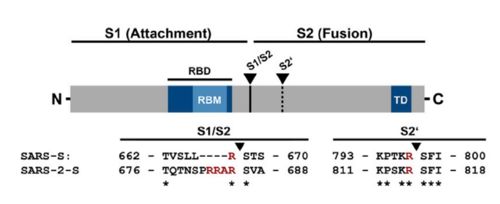 figure 2: Alignment between S protein of SARS and SARS-2 (Hoffmann et al., 2020).
figure 2: Alignment between S protein of SARS and SARS-2 (Hoffmann et al., 2020).


