3D structure representation of the Spike glycoprotein[1] [2].
Introduction
The Spike protein is related to the novel coronavirus pandemic discovered in 2019. The virus is named as the severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) and it belongs to the β coronavirus family. The SARS-COV-2 is a single stranded RNA with 29,881bp length, which encodes for 9860 amino acids. It is compose of two proteins: the structural and non-structural proteins. The structural proteins are S,E,M and N ; whiles as the non-structural proteins are encoded on the ORF regions.
The Spike Protein
The S-protein is a structural protein extends from the viral membrane and it is uniformly arranged as a trimer on the surface to give the crown-like appearance of the SARS-COV-2. The Spike protein is 14-1255bp long, which mediates a receptor binding and fusion of the virus and a cellular membrane. Is also gives the virus its name in Latin as "Corona".
Domains
The S-protein of the SARS-COV-2 has two main subunits , an S1 and S2 subunits. The S1 subunit is located on residue #14–685 ( contains the NTD) and interact with human AEC2 by attaching its virion to the cell membrane by interacting with host receptor. The S2 subunit is located on residue #686–1273 ( contains the CT) which serves as the fusion protein of the virus.
Activation of S-protein
Before the Spike protein can be activated , it has to be cleaved by the protease Furin protein. This 2D image below shows the schematic cleavage of the S-protein before and after it enters the host cell
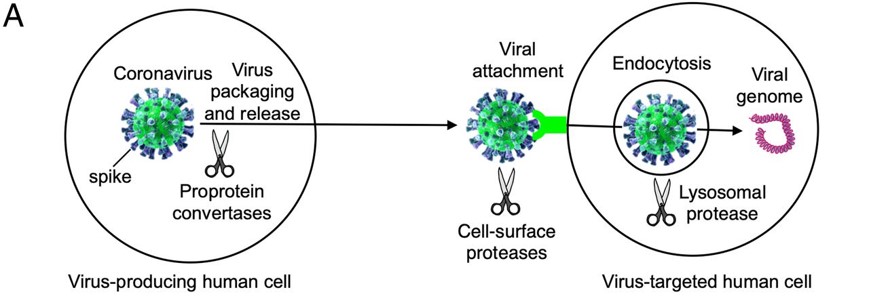
The Mechanism of the Spike Protein
Structural highlights
These are the structural 3D representations of the s-protein showing the two subunits , the binding regions to its receptor human ACE2 , Mutation site , RBD site and cleavage site respectively NTD -CT .Val367 . D614 . LYS 187 .ALA 668

