Introduction
History
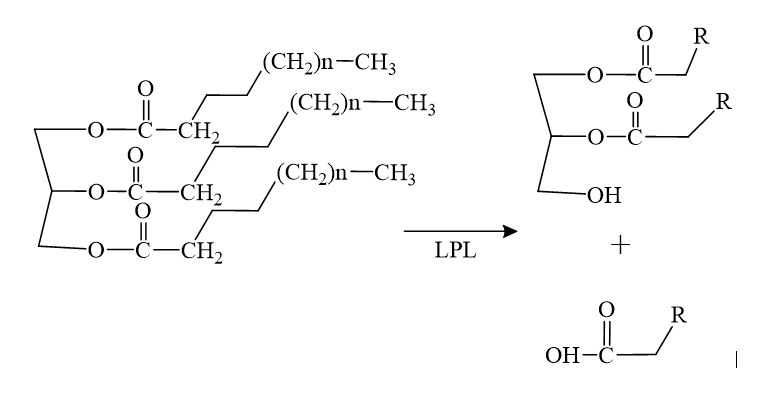
LPL, lipoprotein lipase, is an enzyme that affects the breakdown of triglycerides which are carried from various organs to the blood by molecules called lipoproteins. LPL is found on the surface of cells lining the capillaries within muscles and fatty tissue.
 LPL was identified more than 60 years ago and studied by biochemists and physiologists intensely since. it wasn’t until recently that LPL’s detailed structure was determined due to LPL’s hydrolase domain susceptibility to unfolding. LMF1 and GPIHBP1, glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 ,were discovered to be required for proper folding and enzymatic activity of LPL. LMF1, lipase maturation factor 1, is a chaperone protein that is responsible for proper folding and secretion of LPL. Through the use of X-ray Crystallography it was also discovered that LPL is a monomer rather than the previously believed homodimer.
LPL was identified more than 60 years ago and studied by biochemists and physiologists intensely since. it wasn’t until recently that LPL’s detailed structure was determined due to LPL’s hydrolase domain susceptibility to unfolding. LMF1 and GPIHBP1, glycosylphosphatidylinositol-anchored high density lipoprotein–binding protein 1 ,were discovered to be required for proper folding and enzymatic activity of LPL. LMF1, lipase maturation factor 1, is a chaperone protein that is responsible for proper folding and secretion of LPL. Through the use of X-ray Crystallography it was also discovered that LPL is a monomer rather than the previously believed homodimer.
Function
LPL is located within the interstitial space independently and remains stranded there if not acted upon by GPIHBP1. GPIHBP1 then captures LPL in the interstitial spaces and shuttles it across endothelial cells into the capillary lumen. Once in the capillaries, the LPL-GPIHBP1 complex catalyzes the breakdown of triglycerides in the blood. In doing so, it prevents high levels of triglycerides in the plasma to provide nutrients for vital tissues.
Significance
[1]
Structure
Overall Structure
LPL
GPIHBP1
LPL GPIHBP1 complex
N-terminus of LPL
C-terminus of LPL
Calcium Ion Coordination
Active Site
The of LPL is composed of multiple pieces. The , including residues: W82, V84, W113, Y121, Y158, L160, A185, P187, F212, I221, F239, V260, V264 and K 265, outline the general structure of the active site and provides considerable stability of the active site. The is also an important component of the active site, spanding from residues 243-266, that is vital for the recognition of substrates. The , which is a small portion of the hydrophobic tunnel consisting of residues L160, and W82, aids in the overall stability of the active site and the transition state of substrates catalyzed by the (residues H268, S159, and D183).
Mechanism
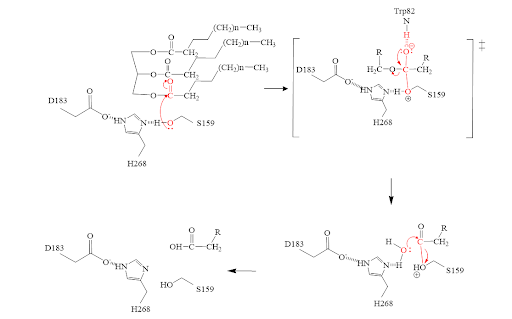
Inhibitors
Disease
Mutations
D201V
M404R
Relevance
Hypertriglycerademia
Drug Relevance


