This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1644
From Proteopedia
| This Sandbox is Reserved from 26/11/2020, through 26/11/2021 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1643 through Sandbox Reserved 1664. |
To get started:
More help: Help:Editing |
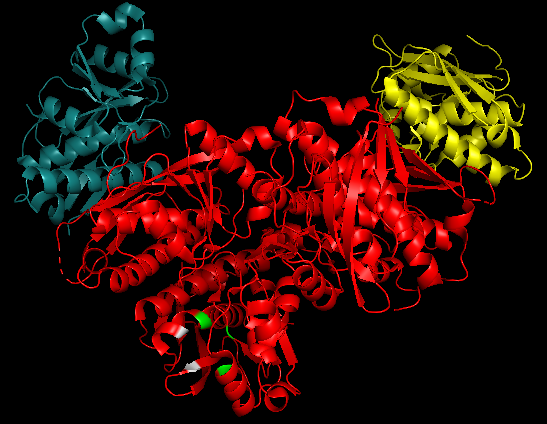
2x36 - Structure of the proteolytic domain of the
| |||||||||||
References
- ↑ Matsushima, Y., Takahashi, K., Yue, S., Fujiyoshi, Y., Yoshioka, H., Aihara, M., ... & Kang, D. (2021). Mitochondrial Lon protease is a gatekeeper for proteins newly imported into the matrix. Communications biology, 4(1), 1-13.
- ↑ Bota, Daniela A., and Kelvin J. A. Davies. “Mitochondrial Lon Protease in Human Disease and Aging: Including an Etiologic Classification of Lon-Related Diseases and Disorders.” Free Radical Biology & Medicine 100 (November 2016): 188–98. https://doi.org/10.1016/j.freeradbiomed.2016.06.031.
- ↑ Bota, Daniela A., and Kelvin J. A. Davies. “Mitochondrial Lon Protease in Human Disease and Aging: Including an Etiologic Classification of Lon-Related Diseases and Disorders.” Free Radical Biology & Medicine 100 (November 2016): 188–98. https://doi.org/10.1016/j.freeradbiomed.2016.06.031.
- ↑ Lu, Bin. “Mitochondrial Lon Protease and Cancer.” Advances in Experimental Medicine and Biology 1038 (2017): 173–82. https://doi.org/10.1007/978-981-10-6674-0_12.
- ↑ Bota, Daniela A., and Kelvin J. A. Davies. “Mitochondrial Lon Protease in Human Disease and Aging: Including an Etiologic Classification of Lon-Related Diseases and Disorders.” Free Radical Biology & Medicine 100 (November 2016): 188–98. https://doi.org/10.1016/j.freeradbiomed.2016.06.031.
- ↑ García-Nafría, Javier, Gabriela Ondrovičová, Elena Blagova, Vladimir M Levdikov, Jacob A Bauer, Carolyn K Suzuki, Eva Kutejová, Anthony J Wilkinson, and Keith S Wilson. “Structure of the Catalytic Domain of the Human Mitochondrial Lon Protease: Proposed Relation of Oligomer Formation and Activity.” Protein Science : A Publication of the Protein Society 19, no. 5 (May 2010): 987–99. https://doi.org/10.1002/pro.376.
- ↑ Lu, Bin. “Mitochondrial Lon Protease and Cancer.” Advances in Experimental Medicine and Biology 1038 (2017): 173–82. https://doi.org/10.1007/978-981-10-6674-0_12.
- ↑ « The N-terminal domain plays a crucial role in the structure of a full-length human mitochondrial Lon protease | Scientific Reports ». Consulté le 13 janvier 2021. https://www.nature.com/articles/srep33631.
- ↑ He, Lihong, Dongyang Luo, Fan Yang, Chunhao Li, Xuegong Zhang, Haiteng Deng, et Jing-Ren Zhang. « Multiple domains of bacterial and human Lon proteases define substrate selectivity ». Emerging Microbes & Infections 7 (17 août 2018). https://doi.org/10.1038/s41426-018-0148-4.
- ↑ Pomatto, L. C., Carney, C., Shen, B., Wong, S., Halaszynski, K., Salomon, M. P., ... & Tower, J. (2017). The mitochondrial Lon protease is required for age-specific and sex-specific adaptation to oxidative stress. Current Biology, 27(1), 1-15.
- ↑ Kutejová, Eva. « Mitochondrial Lon protease-unique structure and essential function in mammalian cells ». Integrative Cancer Science and Therapeutics 5, nᵒ 6 (2018). https://doi.org/10.15761/ICST.1000296.
- ↑ Voos, Wolfgang, et Karen Pollecker. « The Mitochondrial Lon Protease: Novel Functions off the Beaten Track? » Biomolecules 10, nᵒ 2 (7 février 2020). https://doi.org/10.3390/biom10020253.
- ↑ Coscia, F., & Löwe, J. (2021). Cryo‐EM structure of the full‐length Lon protease from Thermus thermophilus. FEBS letters, 595(21), 2691-2700.
- ↑ He, Lihong, Dongyang Luo, Fan Yang, Chunhao Li, Xuegong Zhang, Haiteng Deng, et Jing-Ren Zhang. « Multiple domains of bacterial and human Lon proteases define substrate selectivity ». Emerging Microbes & Infections 7 (17 août 2018). https://doi.org/10.1038/s41426-018-0148-4.
- ↑ Lu, Bin, Swati Yadav, Parul G. Shah, Tong Liu, Bin Tian, Sebastian Pukszta, Nerissa Villaluna, et al. « Roles for the Human ATP-Dependent Lon Protease in Mitochondrial DNA Maintenance ». Journal of Biological Chemistry 282, nᵒ 24 (15 juin 2007): 17363‑74. https://doi.org/10.1074/jbc.M611540200.
- ↑ Kereiche S, Kovacik L, Bednar J, Pevala V, Kunova N, Ondrovicova G, Bauer J, Ambro L, Bellova J, Kutejova E, Raska I. The N-terminal domain plays a crucial role in the structure of a full-length human mitochondrial Lon protease. Sci Rep. 2016 Sep 16;6:33631. doi: 10.1038/srep33631. PMID:27632940 doi:http://dx.doi.org/10.1038/srep33631
- ↑ Garcia-Nafria J, Ondrovicova G, Blagova E, Levdikov VM, Bauer JA, Suzuki CK, Kutejova E, Wilkinson AJ, Wilson KS. Structure of the catalytic domain of the human mitochondrial Lon protease: proposed relation of oligomer formation and activity. Protein Sci. 2010 May;19(5):987-99. PMID:20222013 doi:10.1002/pro.376
- ↑ Garcia-Nafria J, Ondrovicova G, Blagova E, Levdikov VM, Bauer JA, Suzuki CK, Kutejova E, Wilkinson AJ, Wilson KS. Structure of the catalytic domain of the human mitochondrial Lon protease: proposed relation of oligomer formation and activity. Protein Sci. 2010 May;19(5):987-99. PMID:20222013 doi:10.1002/pro.376
- ↑ Garcia-Nafria J, Ondrovicova G, Blagova E, Levdikov VM, Bauer JA, Suzuki CK, Kutejova E, Wilkinson AJ, Wilson KS. Structure of the catalytic domain of the human mitochondrial Lon protease: proposed relation of oligomer formation and activity. Protein Sci. 2010 May;19(5):987-99. PMID:20222013 doi:10.1002/pro.376
- ↑ Wang, N, S Gottesman, M C Willingham, M M Gottesman, and M R Maurizi. “A Human Mitochondrial ATP-Dependent Protease That Is Highly Homologous to Bacterial Lon Protease.” Proceedings of the National Academy of Sciences 90, no. 23 (December 1, 1993): 11247–51. https://doi.org/10.1073/pnas.90.23.11247.
- ↑ Bota, Daniela A., and Kelvin J. A. Davies. “Mitochondrial Lon Protease in Human Disease and Aging: Including an Etiologic Classification of Lon-Related Diseases and Disorders.” Free Radical Biology & Medicine 100 (November 2016): 188–98. https://doi.org/10.1016/j.freeradbiomed.2016.06.031.
- ↑ Bota, Daniela A., and Kelvin J. A. Davies. “Mhttps://proteopedia.org/wiki/skins/common/images/button_extlink.pngitochondrial Lon Protease in Human Disease and Aging: Including an Etiologic Classification of Lon-Related Diseases and Disorders.” Free Radical Biology & Medicine 100 (November 2016): 188–98. https://doi.org/10.1016/j.freeradbiomed.2016.06.031.
- ↑ Bota, Daniela A., and Kelvin J. A. Davies. “Lon Protease Preferentially Degrades Oxidized Mitochondrial Aconitase by an ATP-Stimulated Mechanism.” Nature Cell Biology 4, no. 9 (September 2002): 674–80. https://doi.org/10.1038/ncb836.