Introduction
Neurofibromin (NF) is a cytoplasmic protein encoded by the NF1 gene located on chromosome 17 [1]. It is localized to the cell membrane by the SPRED 1 protein[2]. The neurofibromin protein in encoded by over 350 kb of DNA and contains 62 exons, 58 of which are constitutive[3]. NF suppresses the Ras oncogene by increasing the rate of hydrolysis from (active) to Ras-GDP (inactive)[4]. Increasing the rate of Ras inactivation decreases cell proliferation linked to cancer[5].
Structure
Domains

Figure 1. Domains of Neurofibromin.
GRD domain
The Gap-related domain, or GRD, is the catalytic domain of neurofibromin. This domain also contains a tubulin-binding domain. Its main catalytic mechanism is the hydrolysis of GTP-bound Ras into GDP-bound Ras, which converts Ras from its active form into its inactive form. The GRD provides an arginine residue, known as the arginine finger, to Ras.
SEC-PH
The Sec-PH domain is the lipid-binding domain of neurofibromin. In the of neurofibromin, the hydrophobic core is blocked by the Gap-related domain. The allows the hydrophobic core in the Sec cavity to be accessible and exposed.
CSRD and CTD
The Cysteine-Serine-rich domain (CSRD) and C-terminal domain (CTD) contain phosphorylation sites. The CSRD is able to be phosphorylated by protein kinases A and C. Phosphorylation by protein kinase C is a positive regulator of neurofibromin activity. The CTD is phosphorylated primarily by protein kinase C. This domain is a negative regulator of neurofibromin activity if particular residues are phosphorylated. It also plays an important role in tubulin binding, as it helps in the transition from metaphase to anaphase. CTD contains a nuclear localization signal as well.
Important Structural Features
Conformations
Neurofibromin is a dimeric protein. It exists in two conformations, and . The open conformation has one of the protomers in an auto-inhibited conformation and the other in an open conformation. In the , Ras is able to bind to the GRD neurofibromin. The has both protomers in an autoinhibited conformation, which sterically hinders the binding of Ras to GRD. Only one of the protomers has to be in the open conformation for Ras to bind.
SPRED-1 Protein
The SPRED-1 protein to the cell membrane in to allow it to bind to the membrane oriented Ras protein[6]. SPRED-1 interacts with the N-terminal domain of NF to guide it to the membrane from the cytosol, where its C terminal domain will determine its target [7].
Ras binding site
Ras and Neurofibromin associate through an arginine residue, 1276, that comes from neurofibromin. This arginine is referred to as the “arginine finger” and assists in the hydrolysis of GTP by binding to a backbone carbon atom of tyrosine 32 of Ras when neurofibromin is in the open conformation. It points into the GTP binding site of Ras when neurofibromin is in the open conformation. R1276 also helps stabilize the position of Glutamine 61, a key catalytic residue, through hydrogen bonds.
Glutamine 61 of Ras is a residue that facilitates the conversion of GTP to GDP, turning Ras from its active state to inactive state. There is a catalytic water molecule that glutamine interacts with to position the molecule for a nucleophilic attack on the gamma phosphate of GTP. Mutations of this residue have been related to lower rates of hydrolysis. [8]. Tyrosine 32 makes water-mediated hydrogen bonds with the gamma phosphate of GTP. This position is also where Ras is phosphorylation to promote the activity of GTPase-activating proteins and GTP hydrolysis. [9]
Disease Relevance
Mutations to the neurofibromin protein are implicated in the progression of Neurofibromatosis type 1. Neurofibromatosis type 1 is a condition where cancer develops by inactivating the Ras suppression effects of NF, allowing Ras to behave as an oncogene. Neurofibromatosis type 1 is an autosomal dominant disorder that affects 1 in 3,000 people. The NF gene has the highest mutation rate of any human gene, adding to the prevalence of cancers related to neurofibromin mutations[10]. Furthermore, these mutations consist heavily of de novo mutations[11]. NF1 primarily causes tumors in the central and peripheral nervous systems, but often has a multisystem expression including tumors in the dermatologic, cardiovascular, gastrointestinal, and orthopedic systems[12]. The wide range of presentations is consistent with the multiplicity of mutations observed in the causative protein[13]. This multiplicity derives from the immense size and homo dimeric nature of the neurofibromin protein that allows for otherwise innocuous mutations to wreak havoc on the conformations of the protein as well as its ability to bind to the Ras protein given that the positioning of NF relative to Ras is highly important.
Downstream Effects
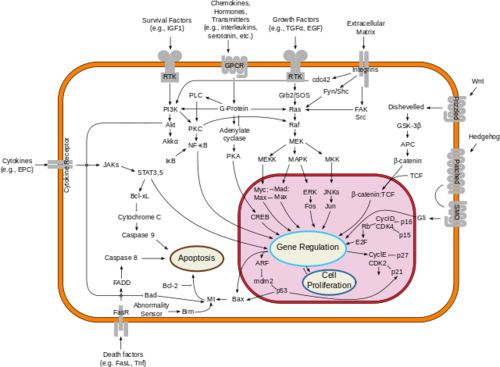
Figure 2: Downstream proliferation of Ras signals have a variety of impacts, including gene regulation, cell proliferation, and cell growth; By cybertory - This file was derived from: Signal transduction v1.png, CC BY-SA 3.0,
https://commons.wikimedia.org/w/index.php?curid=12081090Misregulated Ras activity can lead to uncontrolled signaling in many different cell signaling pathways. Figure 2 provides an overview of the pathways that are connected to Ras. As shown, neurofibromin's ability to regulate Ras activity can alter cell proliferation signaling, activation/ inactivation of transcription factors, and cell apoptotic factors, making its proper functioning crucial [14]. Additionally, due to its importance, neither neurofibromin nor Ras profile as possible drug targets.


