Journal:Proteins:3
From Proteopedia

Do Newly Born orphan proteins resemble Never Born proteins? A study using three deep learning algorithms
Newly Born proteins, or orphan proteins, have no sequence homology to other proteins and occur in single species or within a taxonomically restricted gene family
Never Born proteins are random polypeptides with amino acid content similar to that of native proteins.
Can recently developed AI/Deep Learning tools for predicting 3D protein structures like:
- AlphaFold2 (AF2)
- RoseTTAFold (RTF)
- Evolutionary Scale Modeling (ESM-2)
be useful to see if Newly Born proteins are similar to Never Born proteins? AF2 and RTF predict, by default, five top models, while ESM-2 predicts only one model. Morphing between the top models of AF2 and those of RTF give a visual feeling of how similar these 5 models are for each method.
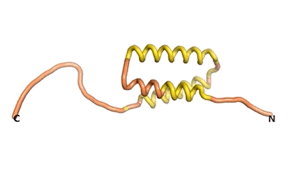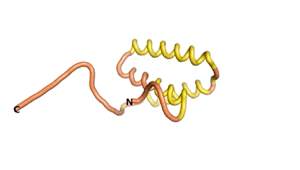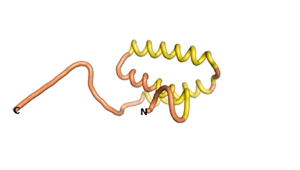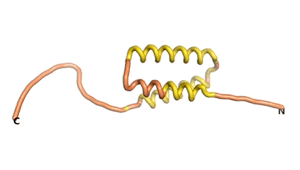
Sequences of the Never Born proteins by Tretyachenko et al.[1] showed that some folded into compact structures, e.g., as seen for Sequences #1856 and #6387.
| RTF-1856 | ESM-1856 | AF2-1856 |
|---|---|---|
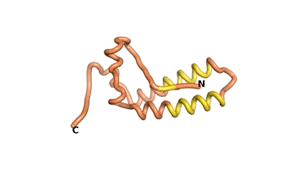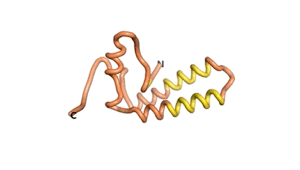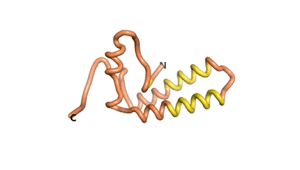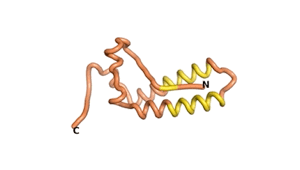
| 
| 
|
| RTF-6387 | ESM-6387 | AF2-6387 |
|---|---|---|
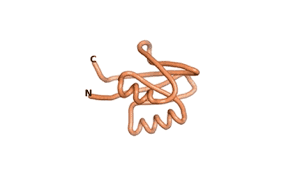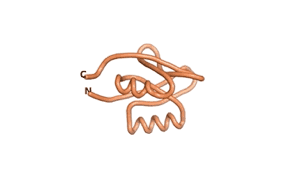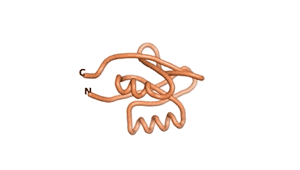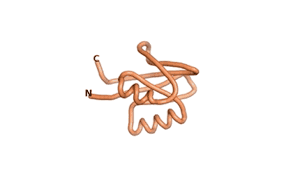
| 
| 
|
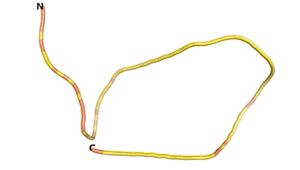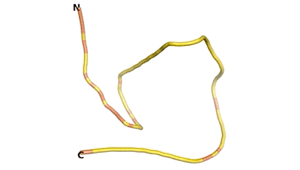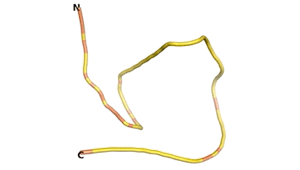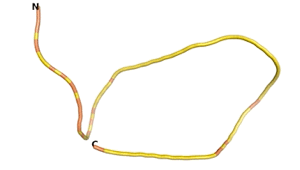
Other never born proteins experimentally appear to belong to the category of intrinsically disordered proteins (IDPs)[2], e.g., as seen for Sequence #3703.
| RTF-3703 | ESM-3703 | AF2-3703 |
|---|---|---|
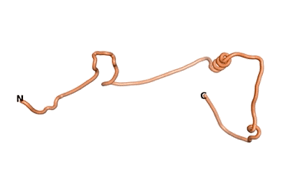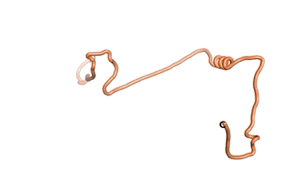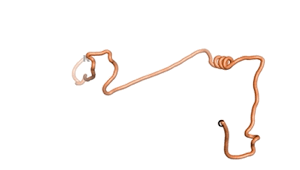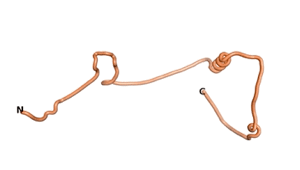
| 
| 
|
References
- ↑ Tretyachenko V, Vymětal J, Bednárová L, Kopecký V Jr, Hofbauerová K, Jindrová H, Hubálek M, Souček R, Konvalinka J, Vondrášek J, Hlouchová K. Random protein sequences can form defined secondary structures and are well-tolerated in vivo. Sci Rep. 2017 Nov 13;7(1):15449. PMID:29133927 doi:10.1038/s41598-017-15635-8
- ↑ Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008 Dec;18(6):756-64. Epub 2008 Nov 17. PMID:18952168 doi:10.1016/j.sbi.2008.10.002
sample
