Journal:Proteins:3
From Proteopedia

Do Newly Born orphan proteins resemble Never Born proteins? A study using three deep learning algorithms
Newly Born proteins, or orphan proteins, have no sequence homology to other proteins and occur in single species or within a taxonomically restricted gene (TRG) family.
Never Born proteins are random polypeptides with amino acid content similar to that of native proteins.
Can recently developed AI/Deep Learning tools for predicting 3D protein structures like:
- AlphaFold2 (AF2)
- RoseTTAFold (RTF)
- Evolutionary Scale Modeling (ESM-2)
be useful to see if Newly Born proteins are similar to Never Born proteins? AF2 and RTF predict, by default, five top models, while ESM-2 predicts only one model. Morphing between the top models of AF2 and those of RTF give a visual feeling of how similar these 5 models are for each method.
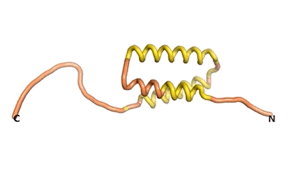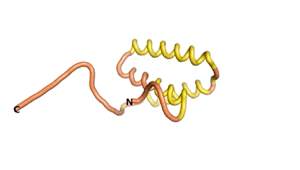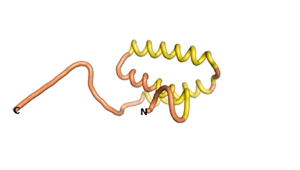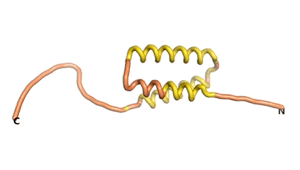
True orphan proteins have no sequence homology to any existing protein. We thought, therefore, that the Never Born proteins generated and investigated by Tretyachenko et al.[1] would serve as a valuable benchmark for comparison. In their study they experimentally showed that some Never Born proteins folded into compact structures, e.g., as seen for Sequences #1856 and #6387.
| RTF-1856 | ESM-1856 | AF2-1856 |
|---|---|---|
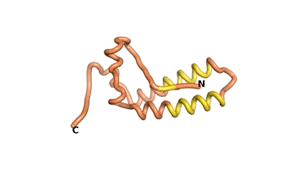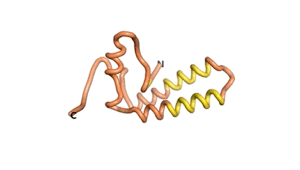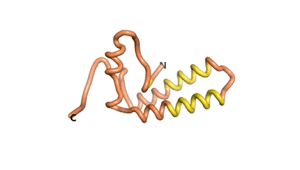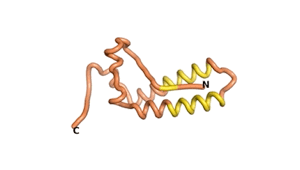
| 
| 
|
| RTF-6387 | ESM-6387 | AF2-6387 |
|---|---|---|
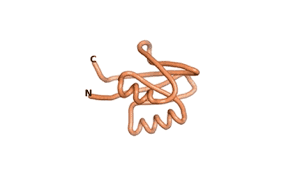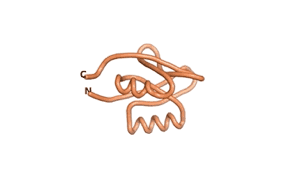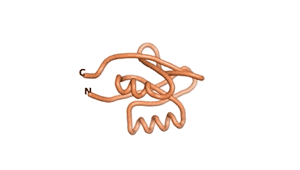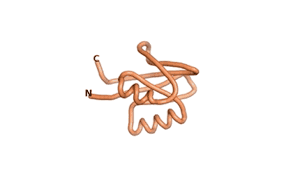
| 
| 
|
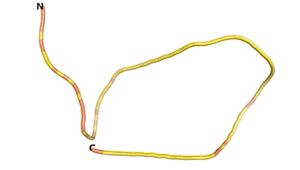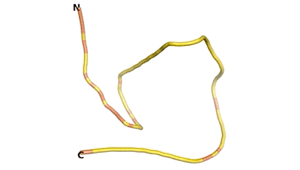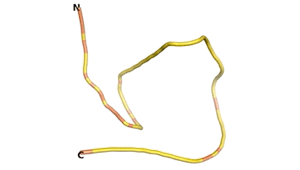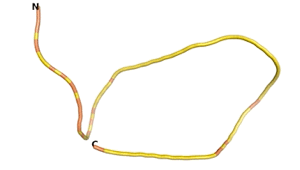
Other never born proteins experimentally appear to belong to the category of intrinsically disordered proteins (IDPs)[2], e.g., as seen for Sequence #3703.
| RTF-3703 | ESM-3703 | AF2-3703 |
|---|---|---|
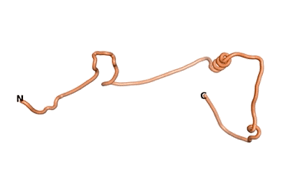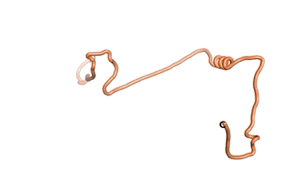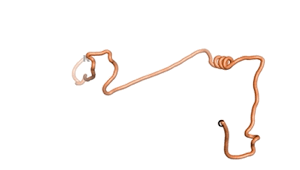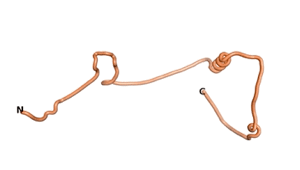
| 
| 
|
We then went on to use the three algorithms on orphan proteins and taxonomically restricted gene products (TRGP) for which no experimental structures were available. We did this in order to see how the predictions of the three algorithms would compare, and whether they would predict novel folds. Although many ORFs have been identified which code for putative orphan proteins, only in a limited number of cases has their association with a well-defined biological activity been established. We have identified seven such proteins for which the necessary sequence data are also available. The number of amino acids for these seven orphans/TRGPs ranges from 109 to 632.
As an initial step in characterizing these seven proteins, we utilized FoldIndex[3] and flDPnn[4] to investigate whether they were predicted to be intrinsically disordered or folded.
References
- ↑ Tretyachenko V, Vymětal J, Bednárová L, Kopecký V Jr, Hofbauerová K, Jindrová H, Hubálek M, Souček R, Konvalinka J, Vondrášek J, Hlouchová K. Random protein sequences can form defined secondary structures and are well-tolerated in vivo. Sci Rep. 2017 Nov 13;7(1):15449. PMID:29133927 doi:10.1038/s41598-017-15635-8
- ↑ Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008 Dec;18(6):756-64. Epub 2008 Nov 17. PMID:18952168 doi:10.1016/j.sbi.2008.10.002
- ↑ Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005 Aug 15;21(16):3435-8. Epub 2005 Jun 14. PMID:15955783 doi:http://dx.doi.org/10.1093/bioinformatics/bti537
- ↑ Hu G, Katuwawala A, Wang K, Wu Z, Ghadermarzi S, Gao J, Kurgan L. flDPnn: Accurate intrinsic disorder prediction with putative propensities of disorder functions. Nat Commun. 2021 Jul 21;12(1):4438. PMID:34290238 doi:10.1038/s41467-021-24773-7
sample
