This is a default text for your page '. Click above on edit this page' to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Function
example/sandbox section
meow
image and green links
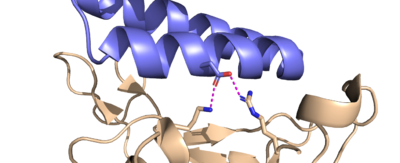
D30 residue
Image of D30 on LCB1 attached to spike
ref
[3]
Reference meow meow meow
[3]
short ref
end
Disease
Relevance
Structural highlights
The goal of designing these minibinders was to create a molecule with a higher binding affinity with the RBD than ACE2, meaning they had to be designed with specific residues that form stronger connections with the same binding pockets that ACE2 would bind to[4]. This section highlights some important residue differences between the minibinders and ACE2 that give the minibinders a higher affinity.
The glutamine-493 (Q493) residue is an important residue in showing the differences in strength between ACE2 and the minibinders[4]. ACE2 doesn’t make use of this residue when binding to the RBD, the nearest residues, Glu-35 and Lys-31 don’t form any interaction with Q493. Comparing this to the AHB2 minibinder, which forms a Hydrogen bond with the Q493 residue, the AHB2 minibinder makes better use of the RBD’s residue than ACE2, helping it have a higher affinity to the spike protein. LCB1 makes even better use of the Q493 residue, forming two hydrogen bonds with two different residues, giving it the highest affinity based on the Q493 residue.
Another important binding site on the RBD includes the Lysine-417 (K417) and Arginine-403 (R403) residues. While ACE2 does form a hydrogen bond interaction with the K417 residue using its own D30 residue, LCB1 forms H bond interactions with both of them, using its own D30 residue, forming a very strong interaction that is hard to break.
It is important to note that these highlighted residues aren’t the only residues that differ between the minibinders, and it is a compilation of all the residue interactions that give each minibinder different affinities. For example, LCB1 forms no interactions with the Q493 residue of the RDB previously mentioned, yet it still has a higher affinity to the RBD than AHB2 which forms a hydrogen bond with the Q493 residue[4].
The goal of designing these minibinders was to create a molecule with a higher binding affinity with the RDB than ACE2, meaning they had to be designed with specific residues that form stronger connections with the same residues that ACE2 would bind to. This section highlights some important residue differences between the minibinders and ACE2 that give the minibinders a higher affinity.
The glutamine-493 (Q493) residue is an important residue in showing the differences in strength between ACE2 and the minibinders. ACE2 doesn’t make use of this residue when binding to the RBD, the nearest residues, Glu-35 and Lys-31 don’t form any interaction with Q493. Comparing this to the AHB2 minibinder, which forms a Hydrogen bond with the Q493 residue, the AHB2 minibinder makes better use of the RDB’s residue than ACE2, helping it have a higher affinity to the spike protein. LCB1 makes even better use of the Q493 residue, forming two hydrogen bonds with two different residues, giving it the highest affinity based on the Q493 residue.
Another important binding site on the RDB includes the Lysine-417 (K417) and Arginine-403 (R403) residues. While ACE2 does form a hydrogen bond interaction with the K417 residue using its own D30 residue, LCB1 forms H bond interactions with both of them, using its own D30 residue, forming a very strong interaction that is hard to break.
It is important to note that these highlighted residues aren’t the only residues that differ between the minibinders, and it is a compilation of all the residue interactions that give each minibinder different affinities. For example, LCB1 forms no interactions with the Q493 residue of the RDB previously mentioned, yet it still has a higher affinity to the RDB than AHB2 which forms a hydrogen bond with the Q493 residue.
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.